|
Size: 4231
Comment:
|
Size: 7066
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| <<BR>> | <<TableOfContents>> |
| Line 9: | Line 9: |
| * Main !BrainStorm window | * Main Brainstorm window |
| Line 20: | Line 20: |
| The tree in the main !BrainStorm window represents the database for the selected study. This database has three levels of definition: Protocol (ie. study, selected in the toolbar), Subject, and Condition. Almost all the operations that can be performed on a file are accessible from the popup menu which is displayed by right clicking on the file. | The tree in the main Brainstorm window represents the database for the selected study. This database has three levels of definition: Protocol (ie. study, selected in the toolbar), Subject, and Condition. Almost all the operations that can be performed on a file are accessible from the popup menu which is displayed by right clicking on the file. |
| Line 28: | Line 28: |
| The following example shows the MEG+EEG protocol "Catching", sorted by condition. There are two experimental conditions, ''Catch ''and ''NoCatch'', and seven subjects. The popup menus shows all the actions that are available for the recordings of subject ''cc'', condition ''Catch''. | The following example shows the MEG+EEG protocol "''Catching''", sorted by conditions. There are two experimental conditions, ''Catch ''and ''!NoCatch'', and seven subjects per condition. The popup menu shows all the actions that are available for the recordings of subject ''cc'', condition ''Catch''. |
| Line 35: | Line 35: |
| Acquisition system: ''EGI GSN - 64 electrodes'' | Acquisition system: ''EGI GSN - Baby 64 electrodes'' |
| Line 48: | Line 48: |
| BrainStorm offers the possibility to reconstruct the cortical activity either on the individual subject anatomy, or on a default anatomy (MNI / Colin27). For this purpose, many interactive tools are available to view, register and process the MR images and the corresponding meshes. However, the cortex segmentation must be performed by external program of your choice ([[Links|list here]]). | Brainstorm offers the possibility to reconstruct the cortical activity either on the individual subject anatomy, or on a default anatomy (MNI / Colin27). For this purpose, many interactive tools are available to view, register and process the MR images and the corresponding meshes. However, the cortex segmentation must be performed by an external program of your choice ([[Links|list here]]). |
| Line 50: | Line 50: |
| Here are a few examples of the views you can obtain with a simple click on the subject's MRI or surface. All the 3D views can be rotated freely with the mouse, zoomed with the wheel, edited with the "Surfaces panel" and with their popup menu. The MRI slices can be moved with a simple mouse operation: right-click and mouse drag. | Here are a few examples of the views you can obtain with a simple click on a MRI or a surface. All the 3D views can be rotated freely with the mouse, zoomed with the wheel, edited with the "Surfaces panel" and with their popup menus. The MRI slices can be moved with a simple mouse operation: right-click and mouse drag. |
| Line 57: | Line 57: |
| All the figures displayed with BrainStorm are always linked in time; and if they represent the same datasets, the sensors selection is also the same for all the views. Selection of a channel of data is done by clicking on it, in a time series or a 3D view. Selected sensors can be displayed separately, marked as "bad", or deleted. | All the figures displayed with Brainstorm are always linked in time; and if they represent the same dataset, the sensors selection is also the same for all the views. The selection of a channel is done by clicking on it, in a time series or a 3D view. Selected channels can be displayed separately, marked as "bad", or deleted. |
| Line 64: | Line 64: |
| Recordings and sources 40Hz low-pass filtering with the "Filters" tab in main BrainStorm window. | Recordings and sources: 40Hz low-pass filtering with the "Filters" tab in main Brainstorm window. |
| Line 71: | Line 71: |
| Acquisition system: ''CTF - 151 sensors'' | Acquisition system: ''CTF MEG - 151 sensors'' |
| Line 73: | Line 73: |
| The ''scouts'' are subsets of the cortical vertices, defined graphically with the "Scouts" tab. They are useful to extract the time series of a the electrical activity of one or several brain regions. | The ''scouts'' are subsets of the cortical vertices, defined graphically with the "''Scouts''" tab. They are useful to extract the time series of a the electrical activity of one or several brain regions. This example shows the cortical response to an electric stimulation of the left index finger. With the two scouts ''Left ''and ''Right'' we can observe the electrical activity in the primary somatosensory cortex in both hemispheres. |
| Line 80: | Line 82: |
| Acquisition system: ''CTF - 151 sensors'' | Acquisition system: ''CTF MEG - 151 sensors'' Same experiment than the previous example, but showing at the same time the responses for condition ''Left-1'' (electric stimulation of the left index) and ''Left-4'' (left ring finger). |
| Line 85: | Line 89: |
== View cortical sources in MRI == Acquisition system: ''CTF MEG - 151 sensors'' Same experiment, with additional display of the scout activity on the 3D MRI slices. [[attachment:snap_scout3D.jpg|{{attachment:snap_scout3D_sm.jpg|attachment:snap_scout3D.jpg}}]] <<BR>> == Time-frequency == Acquisition system: ''CTF MEG - 151 sensors'' Time-frequency decompositions at the sensor or the source level, on small regions of interest or in full resolution. [[attachment:snap_timefreq.jpg|{{attachment:snap_timefreq_sm.jpg|attachment:snap_timefreq.jpg}}]] <<BR>> == Statistical analysis: z-score == Acquisition system: ''EGI GSN - Baby 64 electrodes'' The "''Processes''" tab in the main Brainstorm window allows the user to apply several functions to a set of recordings of sources files. Drag and drop files from the database tree to the white box in the "Processes" tab, and click on "''Run''" to process them. The function that was applied here is a z-score statistic. The algorithm is the following. For each channel: * Compute the mean ''m'' and the variance ''v'' for the baseline * For all the time samples: substract ''m'' and divide by ''v'' The top-right figure represents the initial sources estimate, and the bottom-left figure shows the z-score values for those sources. [[attachment:snap_zscore.jpg|{{attachment:snap_zscore_sm.jpg|attachment:snap_zscore.jpg}}]] <<BR>> == Statistical analysis: t-test == Acquisition system: ''EGI GSN - Baby 64 electrodes'' The "''Processes''" tab can also be used for computing statistical tests, to evaluate the differences between two experimental conditions. This example shows the evaluation of the difference between conditions ''GM'' and ''GMM'', using the results of many different subjects (paired Student t-test, p<0.05). The top figure represents the significance of the difference at the electrodes level across the time, and the bottom figure the thresholded t-values at the cortical level at 280ms. This image also illustrates the "''Coordinates''" tab. It is possible to pick any point from any surface by clicking on it, and immediately get its coordinates in all the coordinates systems used by Brainstorm (MRI, Subject's head and Talairach). [[attachment:snap_ttest.jpg|{{attachment:snap_ttest_sm.jpg|attachment:snap_ttest.jpg}}]] <<BR>> |
Screenshots
Contents
-
Screenshots
- MEG Auditory evoked potential
- Database and right click
- Multiple conditions: Baby auditory EEG
- Subject anatomy: MRI and surfaces
- Channel selection
- Online bandpass filtering
- Cortical region of interest: Scout
- Scouts: multiple conditions
- View cortical sources in MRI
- Time-frequency
- Statistical analysis: z-score
- Statistical analysis: t-test
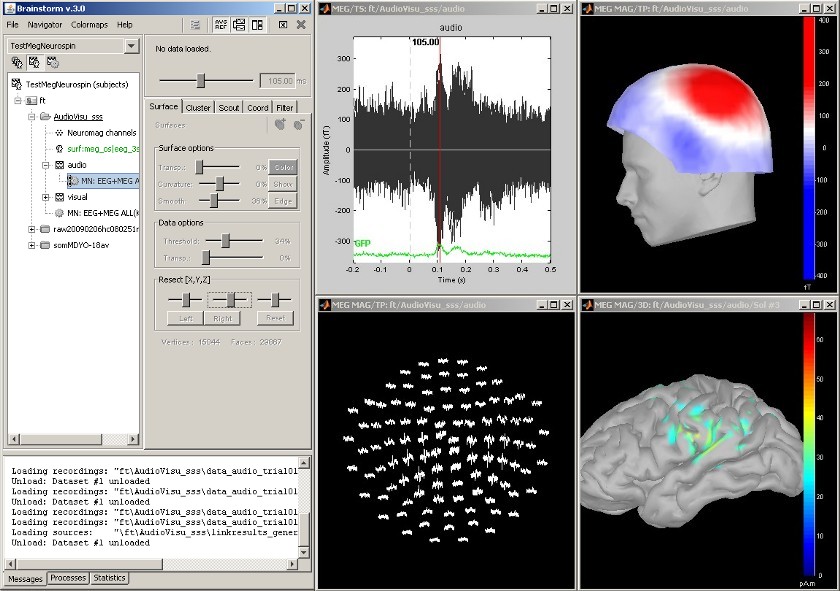
MEG Auditory evoked potential
Acquisition system: Neuromag Vectorview306.
Description of the windows:
- Main Brainstorm window
- Timeseries of all the MEG sensors [-200ms, 500ms]
- Magnetic field recorded by the magnetometers at t=106ms
- Spatial view of the magnetometers time series [-200ms, 500ms]
- Reconstruction of the cortical currents, based on the magnetometers, at t=106ms
Database and right click
The tree in the main Brainstorm window represents the database for the selected study. This database has three levels of definition: Protocol (ie. study, selected in the toolbar), Subject, and Condition. Almost all the operations that can be performed on a file are accessible from the popup menu which is displayed by right clicking on the file.
The first three buttons in the toolbar allows the user to switch between different views of the same database:
- Anatomy: display the MRI and surfaces for each subject
- Functional data (sorted by subject): sensors definition, recordings, sources, statistic results
- Functional data (sorted by condition): idem, but sorted in a different way
The following example shows the MEG+EEG protocol "Catching", sorted by conditions. There are two experimental conditions, Catch and NoCatch, and seven subjects per condition. The popup menu shows all the actions that are available for the recordings of subject cc, condition Catch.
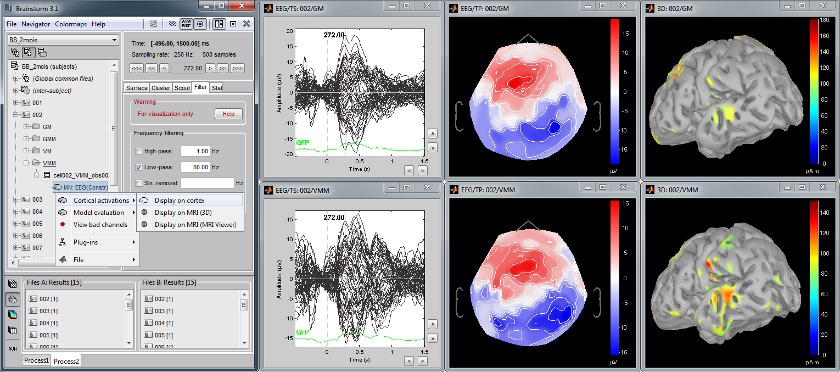
Multiple conditions: Baby auditory EEG
Acquisition system: EGI GSN - Baby 64 electrodes
Description:
- One subject: "001"
- Three conditions: "GM", "GMM", "VM"
- Two views: overlaid electrodes time series, and estimated cortical sources at t=376ms
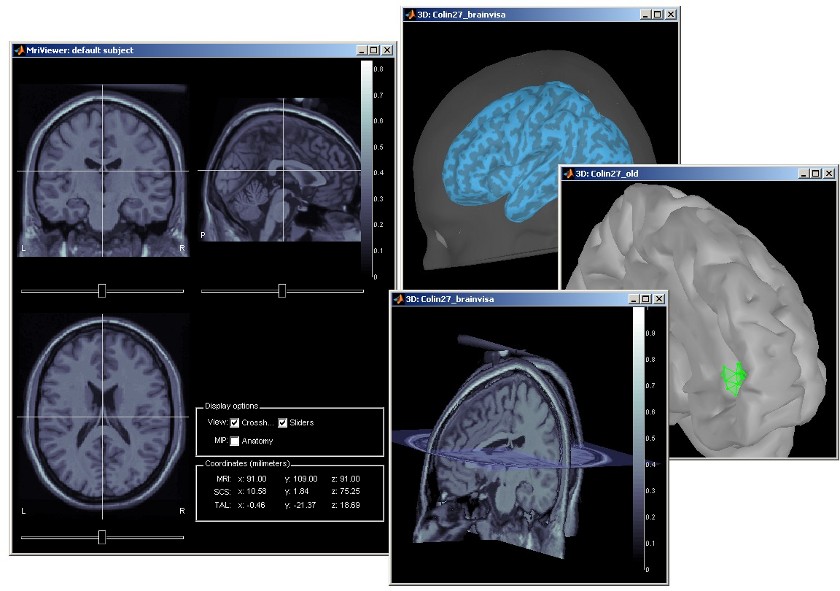
Subject anatomy: MRI and surfaces
Brainstorm offers the possibility to reconstruct the cortical activity either on the individual subject anatomy, or on a default anatomy (MNI / Colin27). For this purpose, many interactive tools are available to view, register and process the MR images and the corresponding meshes. However, the cortex segmentation must be performed by an external program of your choice (?list here).
Here are a few examples of the views you can obtain with a simple click on a MRI or a surface. All the 3D views can be rotated freely with the mouse, zoomed with the wheel, edited with the "Surfaces panel" and with their popup menus. The MRI slices can be moved with a simple mouse operation: right-click and mouse drag.
Channel selection
All the figures displayed with Brainstorm are always linked in time; and if they represent the same dataset, the sensors selection is also the same for all the views. The selection of a channel is done by clicking on it, in a time series or a 3D view. Selected channels can be displayed separately, marked as "bad", or deleted.
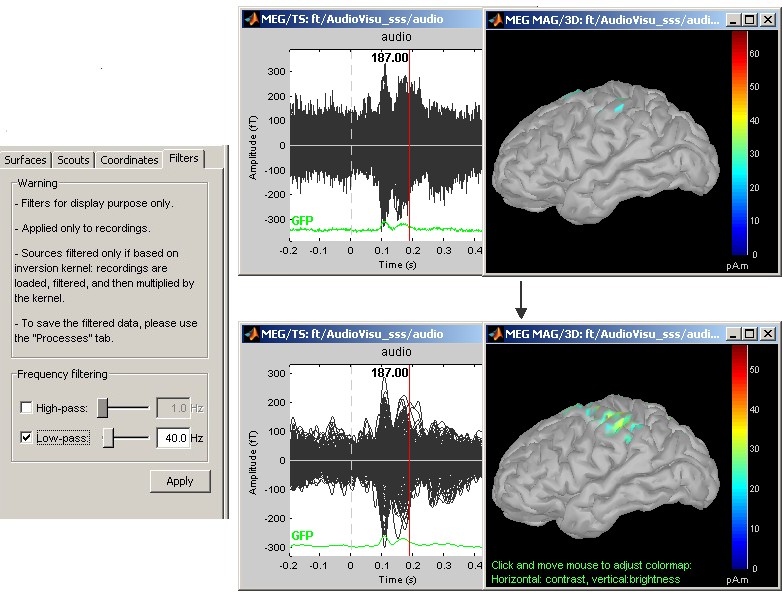
Online bandpass filtering
Recordings and sources: 40Hz low-pass filtering with the "Filters" tab in main Brainstorm window.
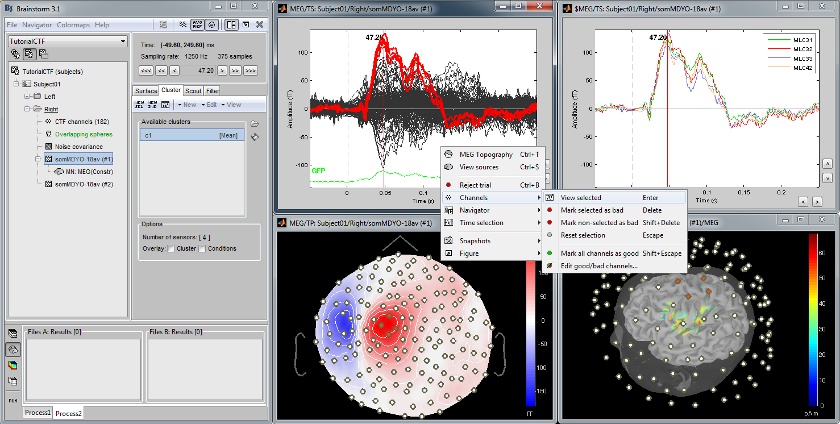
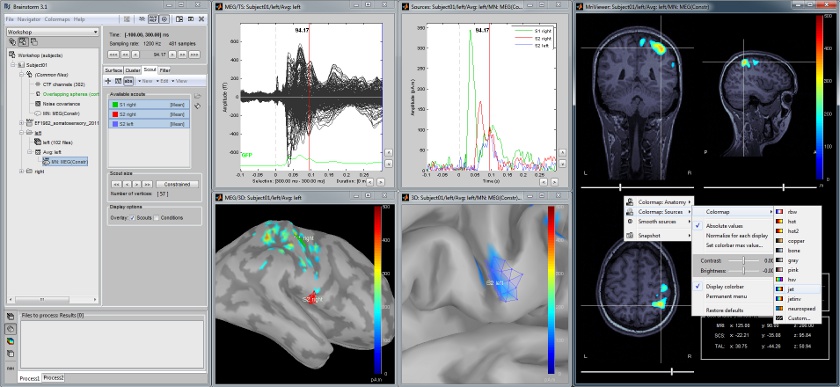
Cortical region of interest: Scout
Acquisition system: CTF MEG - 151 sensors
The scouts are subsets of the cortical vertices, defined graphically with the "Scouts" tab. They are useful to extract the time series of a the electrical activity of one or several brain regions.
This example shows the cortical response to an electric stimulation of the left index finger. With the two scouts Left and Right we can observe the electrical activity in the primary somatosensory cortex in both hemispheres.
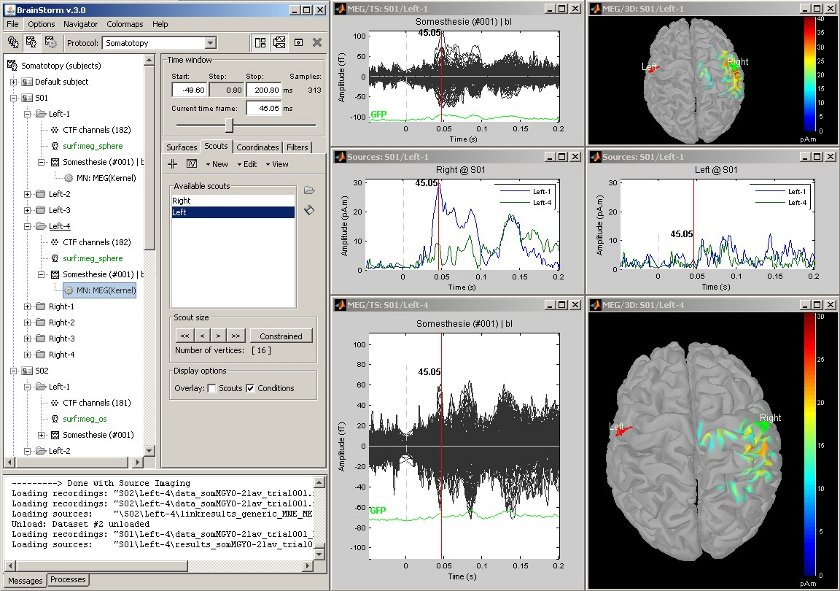
Scouts: multiple conditions
Acquisition system: CTF MEG - 151 sensors
Same experiment than the previous example, but showing at the same time the responses for condition Left-1 (electric stimulation of the left index) and Left-4 (left ring finger).
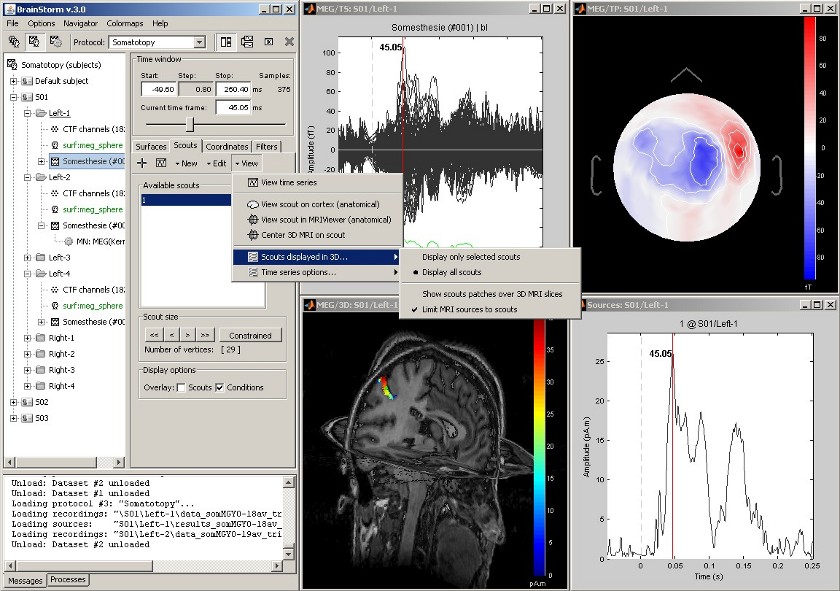
View cortical sources in MRI
Acquisition system: CTF MEG - 151 sensors
Same experiment, with additional display of the scout activity on the 3D MRI slices.
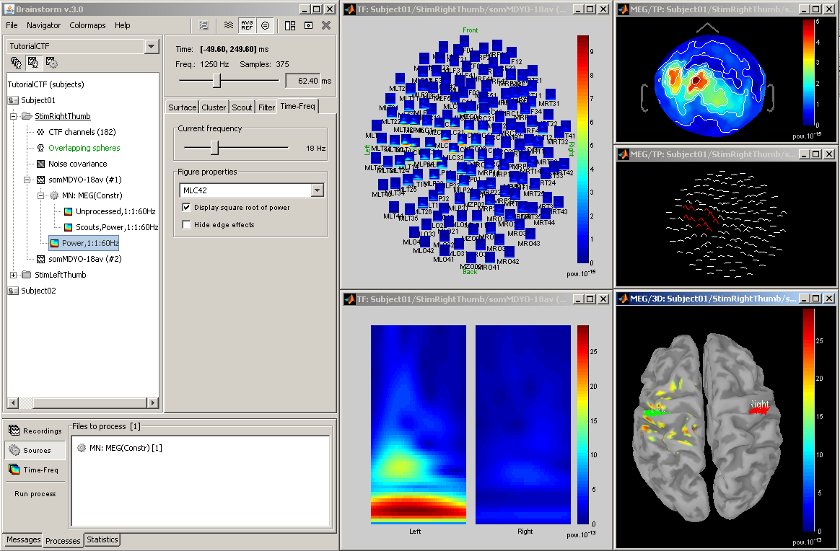
Time-frequency
Acquisition system: CTF MEG - 151 sensors
Time-frequency decompositions at the sensor or the source level, on small regions of interest or in full resolution.
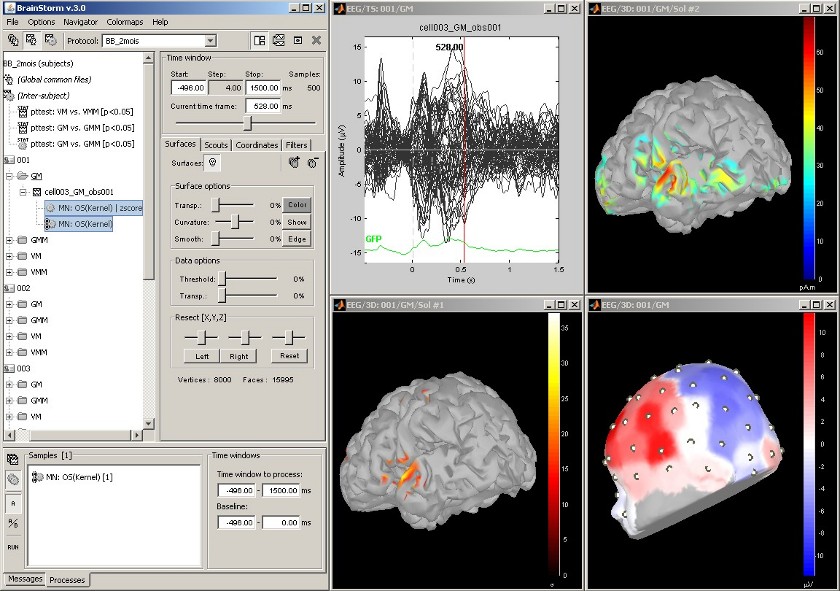
Statistical analysis: z-score
Acquisition system: EGI GSN - Baby 64 electrodes
The "Processes" tab in the main Brainstorm window allows the user to apply several functions to a set of recordings of sources files. Drag and drop files from the database tree to the white box in the "Processes" tab, and click on "Run" to process them.
The function that was applied here is a z-score statistic. The algorithm is the following.
For each channel:
Compute the mean m and the variance v for the baseline
For all the time samples: substract m and divide by v
The top-right figure represents the initial sources estimate, and the bottom-left figure shows the z-score values for those sources.
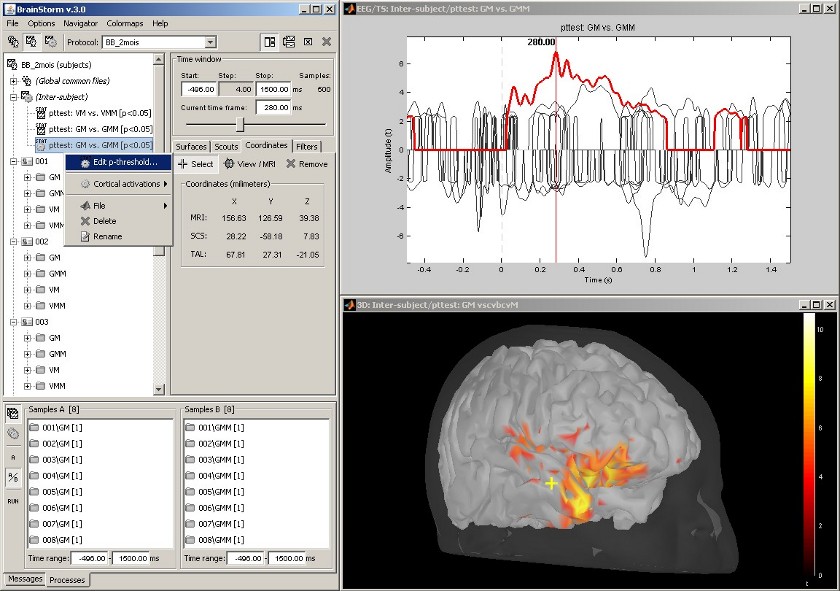
Statistical analysis: t-test
Acquisition system: EGI GSN - Baby 64 electrodes
The "Processes" tab can also be used for computing statistical tests, to evaluate the differences between two experimental conditions.
This example shows the evaluation of the difference between conditions GM and GMM, using the results of many different subjects (paired Student t-test, p<0.05). The top figure represents the significance of the difference at the electrodes level across the time, and the bottom figure the thresholded t-values at the cortical level at 280ms.
This image also illustrates the "Coordinates" tab. It is possible to pick any point from any surface by clicking on it, and immediately get its coordinates in all the coordinates systems used by Brainstorm (MRI, Subject's head and Talairach).