|
Size: 4680
Comment:
|
Size: 5203
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Screenshots = | = Gallery = == Demo video == Tadel F & Baillet S, March 2015<<BR>>Brainstorm: Imaging neural activity at the speed of brain ([[http://neuroimage.usc.edu/videos/201503_brainstorm_demo.mp4|Download video]]) <<HTML(<iframe id="ytplayer" type="text/html" width="853" height="480" src="http://www.youtube.com/embed/30eFJUrRcN4?autoplay=1&origin=http://http://neuroimage.usc.edu/brainstorm" frameborder="0"/>)>> <<HTML(<video width="853" height="480" controls><source src="http://neuroimage.usc.edu/videos/201503_brainstorm_demo.mp4" type="video/mp4"><source src="http://neuroimage.usc.edu/videos/201503_brainstorm_demo.ogg" type="video/ogg">Your browser does not support the video tag.</video>)>> |
| Line 3: | Line 10: |
| Acquisition on an ''CTF 275'' instrument for a left median nerve electric stimulation. <<BR>>[[attachment:snap_median.jpg|{{attachment:snap_median_sm.jpg|attachment:snap_median.jpg}}]] <<BR>> |
Acquisition on a ''CTF 275'' instrument for a left median nerve electric stimulation. <<BR>>[[attachment:snap_median.jpg|{{attachment:snap_median_sm.jpg|attachment:snap_median.jpg}}]] |
| Line 10: | Line 15: |
| <<BR>> == Review continuous recordings and edit markers == |
== Continuous recordings and markers == |
| Line 15: | Line 18: |
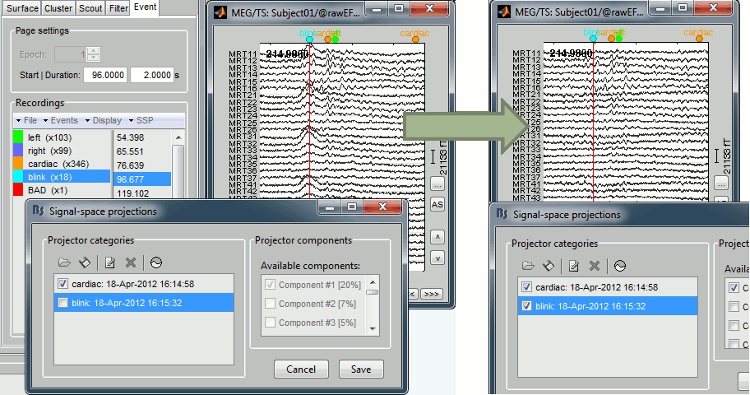
| <<BR>> | == Artifact detection and correction with SSP == A fully automated cleaning pipeline for the most common artifacts (power lines, eye blinks, heartbeats) is available in Brainstorm. The ocular and cardiac artifacts are corrected using the [[Tutorials/TutRawSsp|Signal Space Projection]] approach. === Example: Heartbeat === {{attachment:ssp_ecg.jpg}} === Example: Eye blink === [[attachment:sspExample.gif|{{attachment:sspExample_sm.jpg|attachment:sspExample.gif}}]] |
| Line 20: | Line 30: |
| We provide a few examples of the views you can easily obtain with Brainstorm. All the 3D views can be rotated freely with the mouse, zoomed with the wheel, edited with the "Surface panel" and contextual popup menus. The MRI slices can be browsed with a simple mouse operation: right-click and mouse drag. [[attachment:snap_anatomy.jpg|{{attachment:snap_anatomy_sm.jpg|attachment:snap_anatomy.jpg}}]] <<BR>> |
We provide a few examples of the views you can easily obtain with Brainstorm. All the 3D views can be rotated freely with the mouse, zoomed with the wheel, edited with the "Surface panel" and contextual popup menus. The MRI slices can be browsed with a simple mouse operation: right-click and mouse drag.<<BR>>[[attachment:snap_anatomy.jpg|{{attachment:snap_anatomy_sm.jpg|attachment:snap_anatomy.jpg}}]] |
| Line 27: | Line 33: |
| All the figures displayed by Brainstorm are linked in time. If they feature the same dataset, the sensor selection is also the same for all views. The selection of a channel subset can be easily perfomed by clicking on the corresponding channels in a time series display or a 3D view. Selected channels can be displayed separately, marked as "bad", or deleted. | All the figures displayed by Brainstorm are linked in time. If they feature the same dataset, the sensor selection is also the same for all views. The selection of a channel subset can be easily perfomed by clicking on the corresponding channels in a time series display or a 3D view. Selected channels can be displayed separately, marked as "bad", or deleted.<<BR>>[[attachment:snap_channel.jpg|{{attachment:snap_channel_sm.jpg|attachment:snap_channel.jpg}}]] |
| Line 29: | Line 35: |
| [[attachment:snap_channel.jpg|{{attachment:snap_channel_sm.jpg|attachment:snap_channel.jpg}}]] | == Cortical regions of interest == Acquisition system: ''CTF MEG - 275 sensors'' |
| Line 31: | Line 38: |
| <<BR>> | Scouts are cortical regions of interest, defined graphically from the "Scout" tab. They can be used to extract the time series of MEG and EEG generators within a single or mulitlple brain region. |
| Line 33: | Line 40: |
| == Defining cortical region of interest: Scout == Acquisition system: ''CTF MEG - 151 sensors'' |
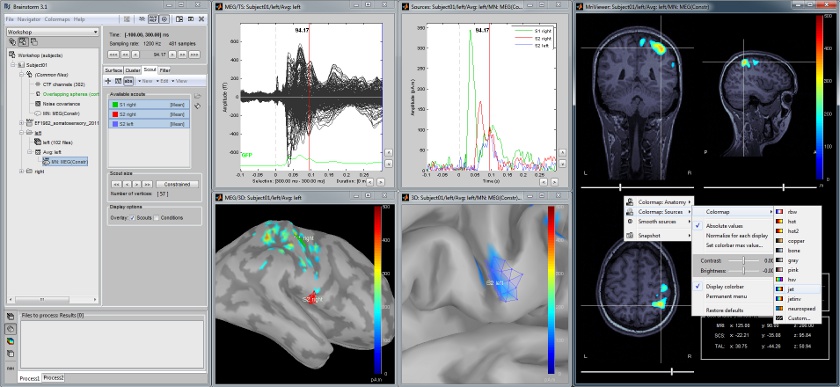
The following example shows the cortical response to an electric stimulation of the left median nerve. With the three scouts "S1 right", "S2 right" and "S2 left"'','' we can track the processing of the stimulus in the brain: 1) contralateral primary somatosensory cortex, 2) contralateral sencondary somatosensory, 3) ipsilateral secondary somatosensory.<<BR>>[[attachment:snap_1scout.jpg|{{attachment:snap_1scout_sm.jpg|attachment:snap_1scout.jpg}}]] |
| Line 36: | Line 42: |
| Scouts are cortical regions of interest, defined graphically from the "Scouts" tab. They can be used to extract the time series of MEG and EEG generators within a single or mulitlple brain region. | == Anatomical atlases == Integrated support for the individual surface-based anatomical atlases generated by FreeSurfer and BrainSuite.<<BR>><<BR>> {{attachment:atlases.jpg||height="358",width="750"}} |
| Line 38: | Line 45: |
| The following example shows the cortical response to an electric stimulation of the left index finger. With the two scouts ''Left ''and ''Right,'' one can observe the electrical activity in the primary somatosensory cortex from each hemisphere. | == Scripting environment == Everything that can be done in the interface with mouse clicks can be converted automatically to Matlab scripts, using the Process1 and Process2 tabs.<<BR>>[[attachment:snap_scripting.jpg|{{attachment:snap_scripting_sm.jpg|attachment:snap_scripting.jpg}}]] |
| Line 40: | Line 48: |
| [[attachment:snap_1scout.jpg|{{attachment:snap_1scout_sm.jpg|attachment:snap_1scout.jpg}}]] | == Digitize and co-register == For accurate source localization in MEG and EEG with registration to anatomical MRI image volume and efficient head localization in MEG, a 3D digitization solution such as the Polhemus-Fastrak is required. Brainstorm features an integrated support for driving a Polhemus device that will help you save time and is efficient for error-checking while digitizing sensor positions and the subject's head shape.<<BR>>The digitized points are used to check and/or fix visually the alignment of the MRI/surfaces and the MEG/EEG sensors.<<BR>><<BR>> {{attachment:polhemus.jpg}} |
| Line 42: | Line 51: |
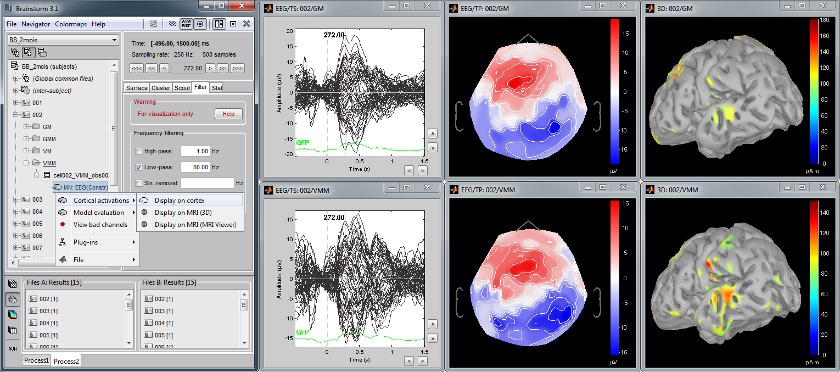
| <<BR>> == From surface to volume: MRI integration == Acquisition system: ''CTF MEG - 151 sensors'' Same experiment as above, with additional 3D display of the scout activity in MRI slices. [[attachment:snap_scout3D.jpg|{{attachment:snap_scout3D_sm.jpg|attachment:snap_scout3D.jpg}}]] <<BR>> == Time-frequency decompositions == Acquisition system: ''CTF MEG - 151 sensors'' Time-frequency decompositions of sensor data and source time series, extracted from cortical regions of interest. [[attachment:snap_timefreq.jpg|{{attachment:snap_timefreq_sm.jpg|attachment:snap_timefreq.jpg}}]] <<BR>> == Statistical inference: t-test maps == Acquisition system: ''EGI GSN - Baby 64 electrodes'' The "''Processes''" tab can also be used for running statistical tests e.g., to evaluate contrasts between two experimental conditions. The following example features the evaluation of the difference between conditions ''GM'' and ''GMM'', from the source maps of mulitple subjects (paired Student t-test, p<0.05). The top figure shows the significance of the difference at the sensor level across time. The bottom figure displays the thresholded t-values at the cortical level at 280ms. This image also illustrates the "''Coordinates''" tab: It is possible to pick any point from any surface by clicking on it, and immediately get its coordinates in all the coordinates systems used by Brainstorm (MRI, Subject's head and Talairach). [[attachment:snap_ttest.jpg|{{attachment:snap_ttest_sm.jpg|attachment:snap_ttest.jpg}}]] <<BR>> Scripting environment Everything that can be done in the interface with mouse clicks can be converted automatically to Matlab scripts, using the Process1 and Process2 tabs. [[attachment:snap_scripting.jpg|{{attachment:snap_scripting_sm.jpg|attachment:snap_scripting.jpg}}]] |
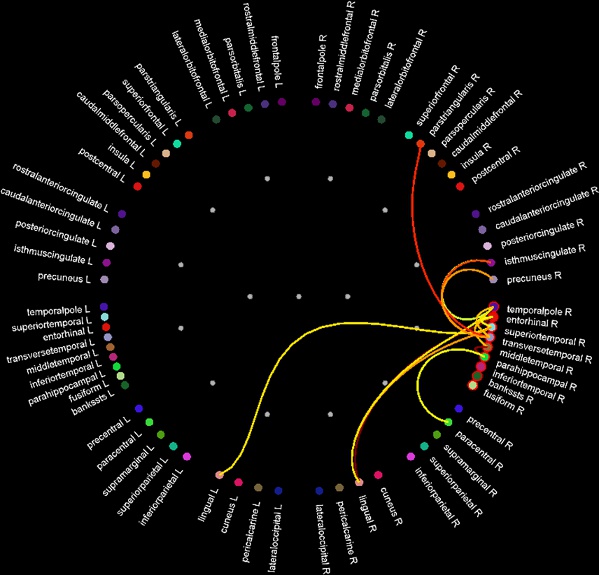
== Functional connectivity == {{attachment:connect_graph.jpg}} |
Gallery
Demo video
Tadel F & Baillet S, March 2015
Brainstorm: Imaging neural activity at the speed of brain (Download video)
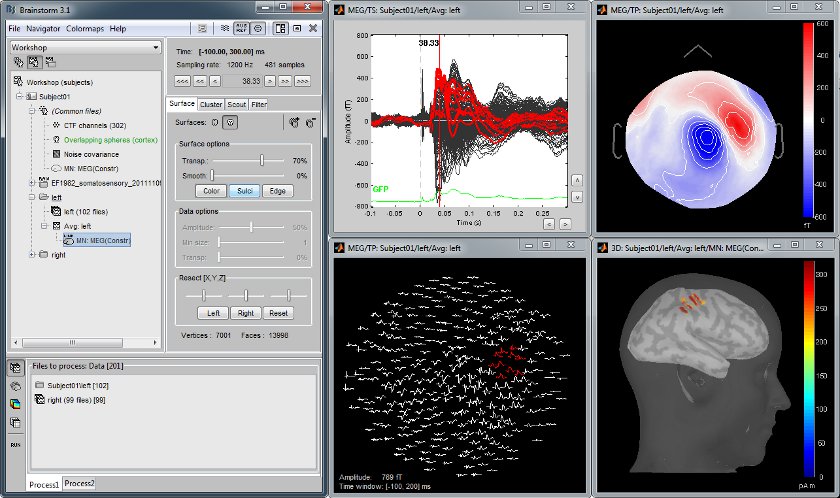
MEG somatosensory evoked responses
Acquisition on a CTF 275 instrument for a left median nerve electric stimulation.
Baby auditory EEG responses
Acquisition system: EGI GSN - Baby 64 electrodes.
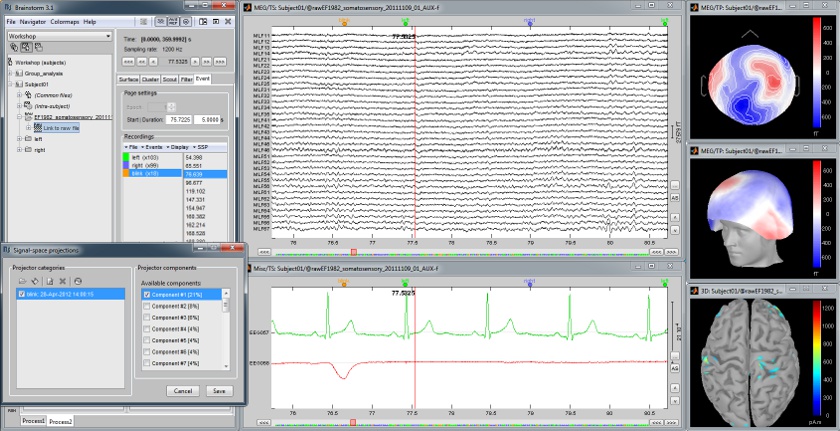
Continuous recordings and markers
Review recordings directly reading from the original files, edit markers, detect and correct artifacts.
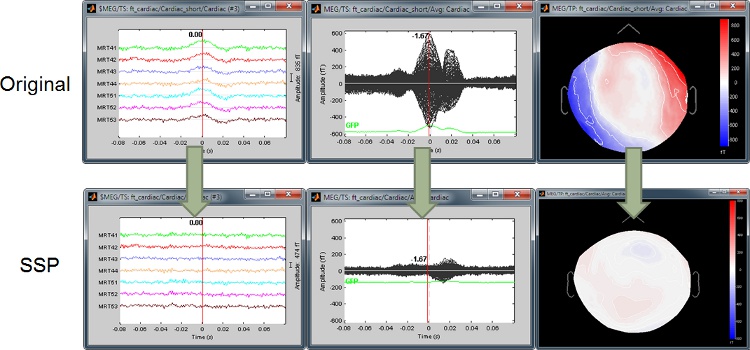
Artifact detection and correction with SSP
A fully automated cleaning pipeline for the most common artifacts (power lines, eye blinks, heartbeats) is available in Brainstorm. The ocular and cardiac artifacts are corrected using the ?Signal Space Projection approach.
Example: Heartbeat
Example: Eye blink
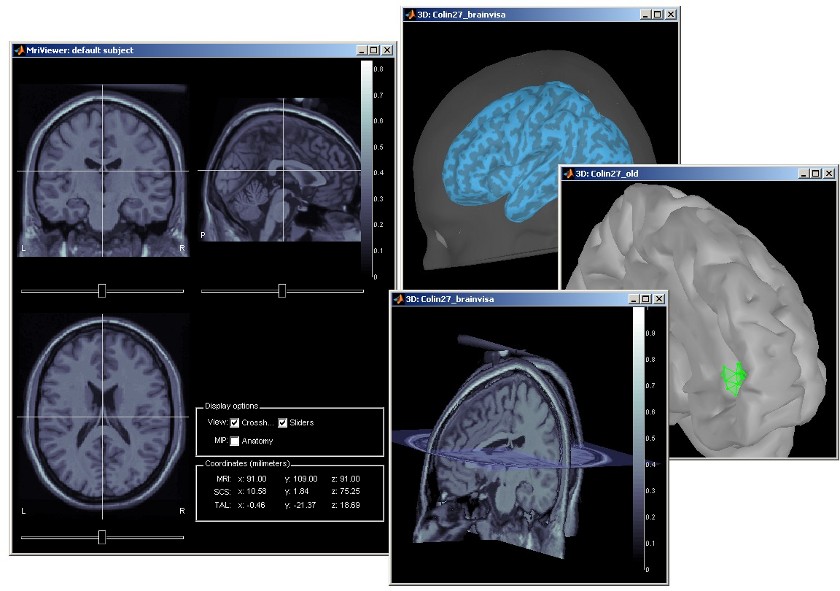
Subject anatomy: MRI and surfaces
Brainstorm features the possibility to model MEG and EEG neural generators either from the individual subject anatomy, or by using a template anatomy (MNI / Colin27) that can be warped to the individual scalp surface. Multiple interactive tools are available to view, register and process the MR images and the corresponding tessellated envelopes. However, tissue segmentation must be performed using another software; multiple options exist today in the academic community (?listed here).
We provide a few examples of the views you can easily obtain with Brainstorm. All the 3D views can be rotated freely with the mouse, zoomed with the wheel, edited with the "Surface panel" and contextual popup menus. The MRI slices can be browsed with a simple mouse operation: right-click and mouse drag.
Channel selection
All the figures displayed by Brainstorm are linked in time. If they feature the same dataset, the sensor selection is also the same for all views. The selection of a channel subset can be easily perfomed by clicking on the corresponding channels in a time series display or a 3D view. Selected channels can be displayed separately, marked as "bad", or deleted.
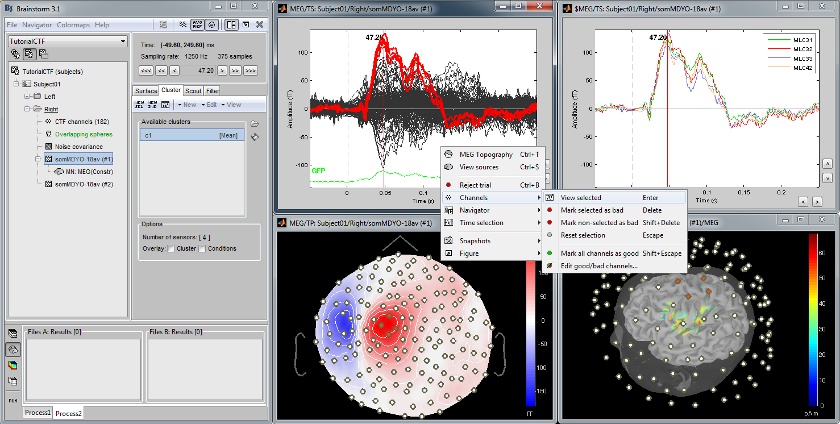
Cortical regions of interest
Acquisition system: CTF MEG - 275 sensors
Scouts are cortical regions of interest, defined graphically from the "Scout" tab. They can be used to extract the time series of MEG and EEG generators within a single or mulitlple brain region.
The following example shows the cortical response to an electric stimulation of the left median nerve. With the three scouts "S1 right", "S2 right" and "S2 left", we can track the processing of the stimulus in the brain: 1) contralateral primary somatosensory cortex, 2) contralateral sencondary somatosensory, 3) ipsilateral secondary somatosensory.
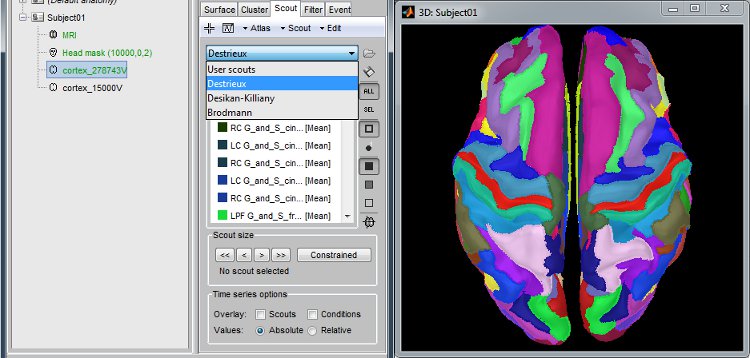
Anatomical atlases
Integrated support for the individual surface-based anatomical atlases generated by FreeSurfer and BrainSuite.
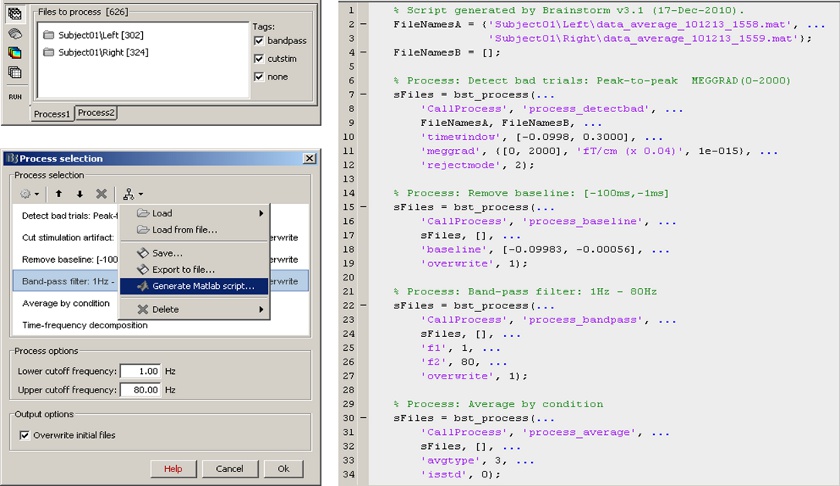
Scripting environment
Everything that can be done in the interface with mouse clicks can be converted automatically to Matlab scripts, using the Process1 and Process2 tabs.
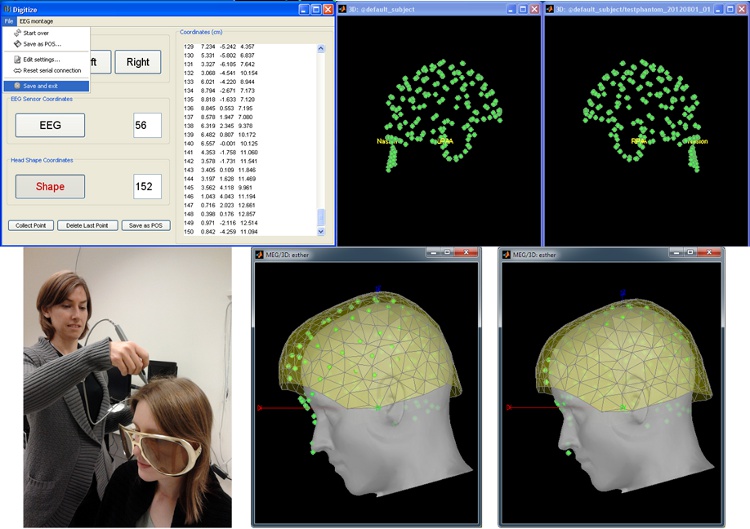
Digitize and co-register
For accurate source localization in MEG and EEG with registration to anatomical MRI image volume and efficient head localization in MEG, a 3D digitization solution such as the Polhemus-Fastrak is required. Brainstorm features an integrated support for driving a Polhemus device that will help you save time and is efficient for error-checking while digitizing sensor positions and the subject's head shape.
The digitized points are used to check and/or fix visually the alignment of the MRI/surfaces and the MEG/EEG sensors.
Functional connectivity