|
Size: 2479
Comment:
|
Size: 3332
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Selecting files in the database = When you have lots of files in a folder, like multiple source reconstructions of time-frequency files for each trial, it is difficult to grab just the ones you are interested in. I improved the process "File > Select files with tag" to help you select only the files you want in the Process tab. '''Example''': You have two source reconstructions with dynamic zscores for each of them, ie. 4 source files per trial: |
= Selecting files in the database [TODO] = ''Authors: Francois Tadel'' When you have lots of files in a folder, like multiple source reconstructions of time-frequency files for each trial, it is difficult to grab just the ones you are interested in. Two mechanisms can help you: the Filter box and the process "File > Select files with tag". ==== Filter text box ==== You want to select only the averages in the data files in the protocol TutorialRaw. Drag and drop all your files there, then use the Filter search box at the bottom-right of the Brainstorm window. Type the string "Avg" to keep only the files that contain this string. {{attachment:allFiles.gif}} The options offered in the Filter menu are: * '''Search comments''': Look for the string in the comments of the files, ie. what is displayed in the database explorer to represent them (the .Comment field) * '''Search file names''': Look for the string in the full file names (including their relative path) * '''Select files''': Only the files that contain the string are selected * '''Exclude files''': Only the files that DO NOT contain the string are selected * '''Reset filters''': Removes the current file filters applied on Process1 and Process2 * '''Case insensitive''': Note that the search is not sensitive to case. If you try to select the constrained sources using the string "Constr" in the comments, it would return all the files, because this matches at the same time "Constr" and "Unconstr". But you can do the opposite instead: exclude the files that contain "Unconstr". ==== Process: Select files with tag ==== You can get to the exact same result by adding the process "File > Select files with tag" before the process you want to execute, to keep only a subset of the files that were placed in the Process1 list. It is less convenient in interactive mode because you don't see immediately the effect of your file filter, but it can be very useful when writing scripts. {{attachment:selectFiles.gif}} To see the list of files that was selected by the process, you can open the report viewer (File > Report viewer). This lists the input files (all the files) and the final files (the selected files), with a summary of how many files where selected on the top. {{attachment:selectReport.gif}} ==== After ==== This process is useless if it is not followed immediately by another process that would do something with the selected files. An application can be the deletion of some files, by adding the process "File > Delete files". ==== How to control the output file names ==== If you are running two processes with different parameters but that produce exactly the same file names and file comments, you wouldn't be able to select them with this process. But immediately after calling any process, you can add the process "File > Add tag" to tag one specific set of files, so that you can re-select them easily later. Example: you run twice the time-frequency decomposition with different options on the same files, tag the files after calculating them with different tags. |
| Line 6: | Line 39: |
| '''Problem''': You just want to select for instance the constrained and z-scored files "MN: MEG(Constr) | abs | zscored". With the tags checkboxes, you can manage to select either all the zscored or non-zscored files, but you cannot separate from the constrained and unconstrained files. '''Solution''': Use the process "File > Select files with tag" to select files with text searches in the file names (what appears on the hard drive, with the ".mat" at the end) or comments (what is displayed in the Brainstorm database explorer). This selects a subset of the files and passes it to the next process in the pipeline. Note that the search is not sensitive to case. If you try to select the constrined sources using the string "Constr" in the comments, it would return all the files, because this matches at the same time "Constr" and "Unconstr". But you can do the opposite instead: exclude the files that contain "Unconstr". '''Results''': To see the list of files that was selected by the process, you can open the report viewer (File > Report viewer). This lists the input files (all the files) and the final files (the selected files), with a summary of how many files where selected on the top. '''After''': This process is useless if it is not followed immediately by another process that would do something with the selected files. An application can be the deletion of some files, by adding the process "File > Delete files". '''How to control the output file names''': If you are running two processes with different parameters but that produce exactly the same file names and file comments, you wouldn't be able to select them with this process. But immediately after calling any process, you can add the process "File > Add tag" to tag one specific set of files, so that you can re-select them easily later. Example: you run twice the time-frequency decomposition with different options on the same files, tag the files after calculating them with different tags. |
== Select files from the pipeline editor [TODO] == * Process: File > Select files: Recordings * Process: File > Select files: Sources * Process: File > Select files: Time-frequency |
Selecting files in the database [TODO]
Authors: Francois Tadel
When you have lots of files in a folder, like multiple source reconstructions of time-frequency files for each trial, it is difficult to grab just the ones you are interested in. Two mechanisms can help you: the Filter box and the process "File > Select files with tag".
Filter text box
You want to select only the averages in the data files in the protocol TutorialRaw. Drag and drop all your files there, then use the Filter search box at the bottom-right of the Brainstorm window. Type the string "Avg" to keep only the files that contain this string.
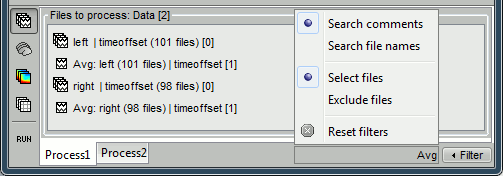
The options offered in the Filter menu are:
Search comments: Look for the string in the comments of the files, ie. what is displayed in the database explorer to represent them (the .Comment field)
Search file names: Look for the string in the full file names (including their relative path)
Select files: Only the files that contain the string are selected
Exclude files: Only the files that DO NOT contain the string are selected
Reset filters: Removes the current file filters applied on Process1 and Process2
Case insensitive: Note that the search is not sensitive to case. If you try to select the constrained sources using the string "Constr" in the comments, it would return all the files, because this matches at the same time "Constr" and "Unconstr". But you can do the opposite instead: exclude the files that contain "Unconstr".
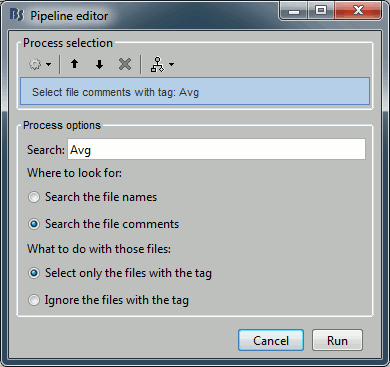
Process: Select files with tag
You can get to the exact same result by adding the process "File > Select files with tag" before the process you want to execute, to keep only a subset of the files that were placed in the Process1 list. It is less convenient in interactive mode because you don't see immediately the effect of your file filter, but it can be very useful when writing scripts.
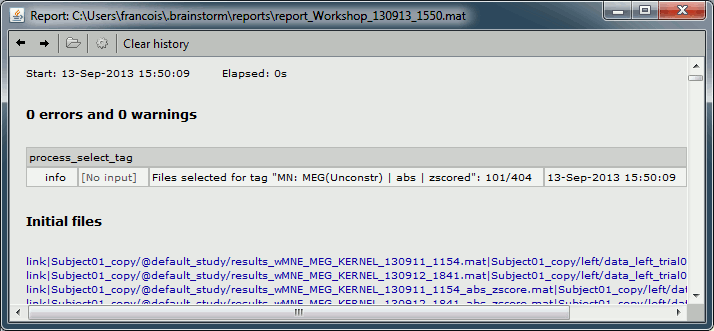
To see the list of files that was selected by the process, you can open the report viewer (File > Report viewer). This lists the input files (all the files) and the final files (the selected files), with a summary of how many files where selected on the top.
After
This process is useless if it is not followed immediately by another process that would do something with the selected files. An application can be the deletion of some files, by adding the process "File > Delete files".
How to control the output file names
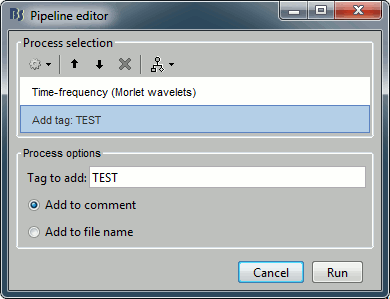
If you are running two processes with different parameters but that produce exactly the same file names and file comments, you wouldn't be able to select them with this process. But immediately after calling any process, you can add the process "File > Add tag" to tag one specific set of files, so that you can re-select them easily later.
Example: you run twice the time-frequency decomposition with different options on the same files, tag the files after calculating them with different tags.
Select files from the pipeline editor [TODO]
Process: File > Select files: Recordings
Process: File > Select files: Sources
Process: File > Select files: Time-frequency
