Brain-fingerprinting
Authors: Jason da Silva Castanheira, Raymundo Cassani
This tutorial introduces the concept of neurophysiological brain-fingerprinting and its implementation in Brainstorm. To follow this tutorial, please first complete the OMEGA tutorial as we will derive the brain-fingerprints of its five participants.
Introduction
Brain-fingerprinting is a method to assess inter-individual differences in brain activity. It was first proposed by Finn and colleagues in 2015. This tutorial will derive spectral brain-fingerprints using neural power spectra at computed within each parcel of the Destrieux atlas. Once the individuals' brain-fingerprints have been obtained, differentiation accuracy and differentiability are computed for the cohort. Finally, we analyze relative contribution of each of the cortical parcels (i.e., brain regions) and frequency bins to participant differentiation
This demonstration uses the pre-processed data from five participants obtained from the outputs of the OMEGA tutorial. Please run the entire tutorial before using the tutorial_brain_fingerprint.m script (shown at the end of this page).
Preparing the data for brain-fingerprinting
The brain-fingerprint of an individual can be derived from functional connectomes or neural power spectrum, as detailed in da Silva Castanheira et al., 2021. Note that this is not an exhaustive list of all possible neurophysiological features that can be used to define brain-fingerprints.
From the estimated surface source maps, we compute the power spectrum density (i.e., PSD) using the Welch method at every vertex of the cortical surface, and average the resulting spectra within each region defined by the Destrieux atlas (see Scout page for more details). Note, this is done to downsample the spatial features of the brain-fingerprint. If we did not downsample the PSD to a cortical atlas, the resulting feature space would be too large to work with (e.g., 15,000 vertices × 300 frequency bins). The choice of atlas and neurophysiological features (e.g., functional connectomes vs power spectra) to define the brain-fingerprint is dependent on the user's specific hypotheses. See the section of the script entitled Compute PSD for all ROIs of an atlas
Brain-fingerprinting requires at least two data segments for every individual. This can be two separate recordings (i.e., between-session fingerprinting), or two segments within the same recording session (i.e., within-session fingerprinting). For this demonstration, we will split the MEG recordings of the OMEGA tutorial into two parts (data segments). As such, for each participant, there will be two brain-fingerprints (b-fp1 and b-fp2), each consisting of a vector of nParcellations×nFrequencies elements. See the image below for a summary of the pipeline:
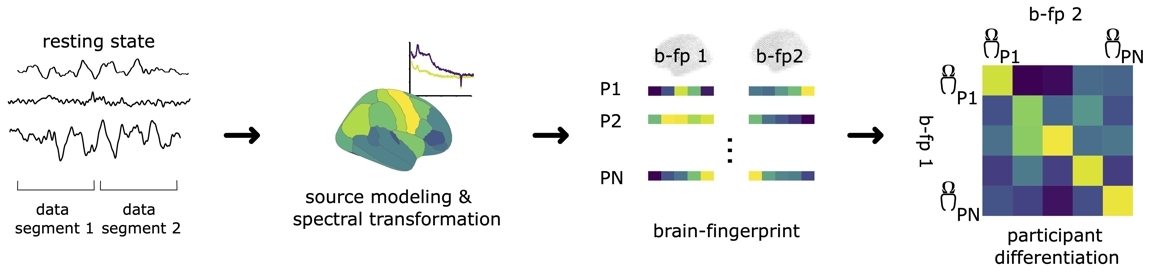
The brain-fingerprinting method
The brain-fingerprinting method is based on the correlational similarity of participants across data segments (see image above). For each participant in a given cohort, we compute the Pearson correlation coefficient between the brain-fingerprint of the first segment and second segment of all participants in the cohort, including the probe participant. This yields a participant similarity matrix, where off-diagonal elements represent the similarity of a participant to all other participants and diagonal elements represent the similarity of a participant's brain-fingerprint across data segments (i.e., similarity to their own brain-fingerprint). See the image below for an example participant similarity matrix.
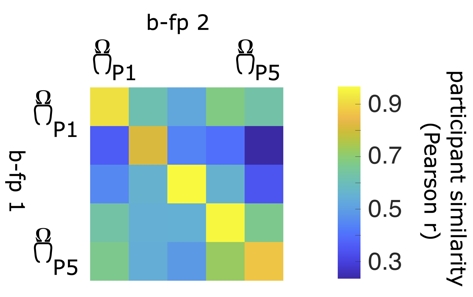
Participant differentiation consists of a lookup procedure along the rows (or columns) of the participant similarity matrix. A participant is said to be correctly differentiated if a participant's brain-fingerprint is more similar to themselves (self-similarity, also referred to as Iself) than all other participants in the cohort (other-similarity, also referred to as Iothers). In other words, if the brain-fingerprint of participant i is most similar to their brain-fingerprint taken at the second time point, this participant is said to be correctly differentiated.
Differentiation accuracy
Differentiation accuracy represents the percent ratio of the number of participants correctly differentiated across the cohort (i.e., differentiation accuracy). Brain-fingerprinting is typically repeated for all possible pairs of data segments. In the case of this tutorial, there are only two data segments, the first and second half of the recording. We therefore obtain two differentiation accuracies along the rows and columns of the participant similarity matrix respectively. In the case where three data segments are available, we can compute six differentiation accuracies: the accuracy for the columns and rows of data segment 1 vs segment 2, and for each possible pair of recordings (i.e., data segment 2 vs 3 and data segment 1 vs 3).
Differentiability
Differentiability is a measure which describes how easily a given participant is to differentiate from a cohort of individuals. This measure consists of scaling (z-scoring) the self-similarity of brain-fingerprints against the other-similarity (off-diagonal elements). A participant with a high differentiability score will have a higher self-similarity relative to their similarity to others in the cohort (other-similarity) and therefore are easily differentiated (see the last row in the example participant similarity matrix below). In contrast, a participant whose brain-fingerprint is most similar to others in the cohort will be more challenging to differentiate (see the second row of the participant similarity matrix below). 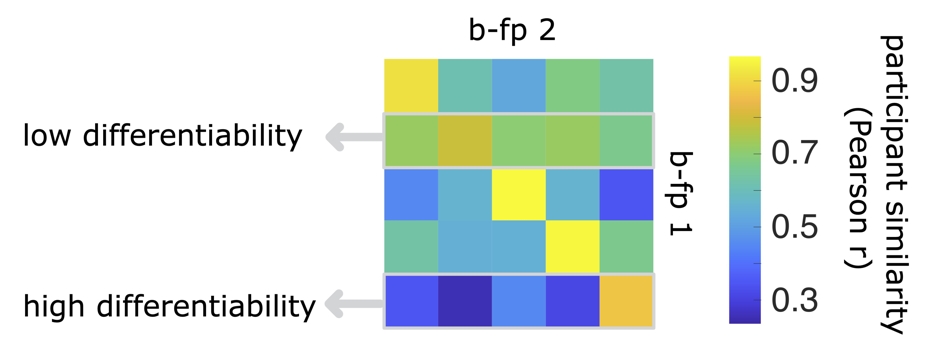
Note that other measures to quantify the relative easy of differentiating a participant exist: Idiff represents the difference between Iself and Iothers. See Amico & Goñi 2018 and Sareen at el., 2021.
The participant similarity matrix is generally symmetric (although it does not necessarily have to be). This implies that the differentiability across rows and columns are generally strongly correlated, as such, the participant differentiability can be computed as the mean of these two per participant
Quantifying the most salient features of the brain-fingerprint
Beyond differentiation accuracy and differentiability, we can quantify the relative contribution of cortical parcels (i.e., brain regions) and frequency bins to participant differentiation.
To do so, we calculated intraclass correlations (ICC). ICC quantifies the ratio of within-participant variance and between-participant variance, with participants as their own raters across data segments (see Amico et al., 2018 & da Silva Castanheira et al., 2021 for details). Features that contribute the most to participant differentiation should be highly consistent within individuals, but show great inter-individual variance. Larger values of ICC indicate that a neurophysiological feature (e.g., region of interest or frequency band) contributes more to participant differentiation than smaller values.
See the subsection entitled IntraClass Correlations (ICC) under the section SIMILARITY AND DIFFERENTIABILITY in the script.
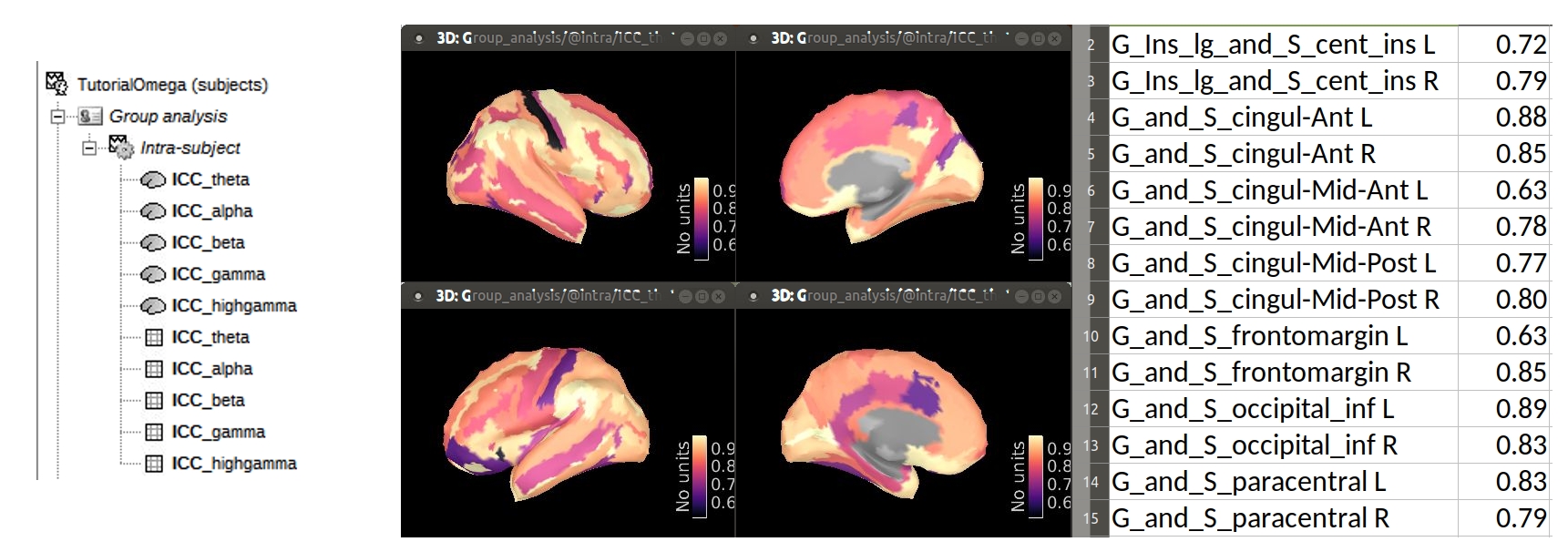
In this tutorial we plot the average ICC value per cortical parcel within each frequency band of interest as defined in the script. See image below:
% Bands for ICC cortex plot
BandNames = {'theta', 'alpha', 'beta', 'gamma', 'highgamma'};
BandLowerFreqs = [ 4, 8, 13, 30, 50]; % Hz, inclusive
BandUpperFreqs = [ 8, 13, 30, 50, 150]; % Hz, exclusiveIn the Brainstorm database, the ICC values are saved: as (i) Matrix files (which can be exported to tabular files) and, as (ii) cortical maps (which allow to visualize their spatial distribution). Both set of files are found in the Group analysis folder. The image below presents the database (left), and ICC values for theta band, as cortical maps (center), and as matrix (right).
Applications of the brain-fingerprint
Beyond biometric differentiation, brain-fingerprints advance the neurobiological origins of individual traits and inform clinical. Below we summarize some of the most recent applications of brain-fingerprints.
Decoding individual traits: Previous work demonstrates that beyond biometric identification, brain-fingerprints can predict individual differences in intelligence test performance (e.g., Finn et al., 2015; Amico et al., 2018) and demographics (da Silva Castanheira et al., 2021).
Clinical application: Recent work has explored the clinical utility of bran-fingerprints. This work has shown across multiple neurological diseases that the brain-fingerprints of patients show increased variability (lower self-similairty/ Iself ) in brain-fingerprints of clinical populations (e.g., Kaufmann et al., 2017, 2018; Sorrentino et al., 2021; Troisi Lopez et al., 2023; da Silva Castanheira et al., 2024). A potential explanation for the de-individualization of brain activity in observed in Parkinson’s disease may be an inherent instability in the arrhythmic brain activity of patients over short periods. In contrast, rhythmic brain activity appears particularly characteristic of patients with PD (da Silva Castanheira et al., 2024). Future work is exploring the application of brain-fingerprints to the design of personalized medicine.
Additional documentation
Please cite previous work on brain-fingerprinting:
da Silva Castanheira, J., Orozco Perez, H.D., Misic, B. et al. Brief segments of neurophysiological activity enable individual differentiation. Nat Commun 12, 5713 (2021). https://doi.org/10.1038/s41467-021-25895-8
Finn, E., Shen, X., Scheinost, D. et al. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci 18, 1664–1671 (2015). https://doi.org/10.1038/nn.4135
Further reading:
Amico, E., Goñi, J. The quest for identifiability in human functional connectomes. Sci Rep (2018). https://doi.org/10.1038/s41598-018-25089-1
Sareen, E., Zahar, S., Ville, D. V. D., Gupta, A., Griffa, A., & Amico, E. Exploring MEG brain fingerprints: Evaluation, pitfalls, and interpretations. NeuroImage (2021). https://doi.org/10.1016/j.neuroimage.2021.118331
Sorrentino, P., Rucco, R., Lardone, A., Liparoti, M., Troisi Lopez, E., Cavaliere, C., Soricelli, A., Jirsa, V., Sorrentino, G., & Amico, E. Clinical connectome fingerprints of cognitive decline. NeuroImage, (2021). https://doi.org/10.1016/j.neuroimage.2021.118253
Troisi Lopez, E., Minino, R., Liparoti, M., Polverino, A., Romano, A., De Micco, R., Lucidi, F., Tessitore, A., Amico, E., Sorrentino, G., Jirsa, V., & Sorrentino, P. Fading of brain network fingerprint in Parkinson’s disease predicts motor clinical impairment. Human Brain Mapping, (2023). https://doi.org/10.1002/hbm.26156
Kaufmann, T., Alnæs, D., Brandt, C. L., Bettella, F., Djurovic, S., Andreassen, O. A., & Westlye, L. T. Stability of the Brain Functional Connectome Fingerprint in Individuals With Schizophrenia. JAMA Psychiatry, (2018). https://doi.org/10.1001/jamapsychiatry.2018.0844
Kaufmann, T., Alnæs, D., Doan, N. T., Brandt, C. L., Andreassen, O. A., & Westlye, L. T. Delayed stabilization and individualization in connectome development are related to psychiatric disorders. Nat Neurosci, (2017). https://doi.org/10.1038/nn.4511
da Silva Castanheira, J., Wiesman, A. I., Hansen, J. Y., Misic, B., Baillet, S., Network Quebec Parkinson, & PREVENT-AD Research Group. (2023). Neurophysiological brain-fingerprints of motor and cognitive decline in Parkinson’s disease. medRxiv.
Tutorials
Scripting
The following script from the Brainstorm distribution reproduces the analysis presented in this tutorial page: brainstorm3/toolbox/script/tutorial_brain_fingerprint.m
1 function tutorial_brain_fingerprint(ProtocolNameOmega, reports_dir)
2 % TUTORIAL_BRAIN_FINGERPRINT: Script that reproduces the results of the online tutorial "Brain-fingerprint".
3 %
4 % CORRESPONDING ONLINE TUTORIAL:
5 % https://neuroimage.usc.edu/brainstorm/Tutorials/BrainFingerprint
6 %
7 % INPUTS:
8 % - ProtocolNameOmega : Name of the protocol created with all the data imported (TutorialOmega)
9 % - reports_dir : Directory where to save the execution report (instead of displaying it)
10 %
11 % @=============================================================================
12 % This function is part of the Brainstorm software:
13 % https://neuroimage.usc.edu/brainstorm
14 %
15 % Copyright (c) University of Southern California & McGill University
16 % This software is distributed under the terms of the GNU General Public License
17 % as published by the Free Software Foundation. Further details on the GPLv3
18 % license can be found at http://www.gnu.org/copyleft/gpl.html.
19 %
20 % FOR RESEARCH PURPOSES ONLY. THE SOFTWARE IS PROVIDED "AS IS," AND THE
21 % UNIVERSITY OF SOUTHERN CALIFORNIA AND ITS COLLABORATORS DO NOT MAKE ANY
22 % WARRANTY, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO WARRANTIES OF
23 % MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE, NOR DO THEY ASSUME ANY
24 % LIABILITY OR RESPONSIBILITY FOR THE USE OF THIS SOFTWARE.
25 %
26 % For more information type "brainstorm license" at command prompt.
27 % =============================================================================@
28 %
29 % Author: Jason da Silva Castanheira, Raymundo Cassani, 2024
30
31
32 %% ===== CHECK PROTOCOL =====
33 % Start brainstorm without the GUI
34 if ~brainstorm('status')
35 brainstorm nogui
36 end
37 % Output folder for reports
38 if (nargin < 2) || isempty(reports_dir) || ~isdir(reports_dir)
39 reports_dir = [];
40 end
41 % You have to specify the folder in which the tutorial dataset is unzipped
42 if (nargin < 1) || isempty(ProtocolNameOmega)
43 ProtocolNameOmega = 'TutorialOmega';
44 end
45
46
47 %% ===== BRAIN-FINGERPRINT PARAMETERS =====
48 % Subjects
49 SubjectNames = {'sub-0002', 'sub-0003', 'sub-0004', 'sub-0006', 'sub-0007'};
50 nSubjects = length(SubjectNames);
51 % Frequency range
52 LowerFreq = 4; % Hz, inclusive
53 UpperFreq = 150; % Hz, exclusive
54 % Cortical parcellation (atlas)
55 % Note we downsample the cortical surface using an atlas for computation efficiency
56 AtlasName = 'Destrieux';
57 AtlasScouts = {'G_Ins_lg_and_S_cent_ins L', 'G_Ins_lg_and_S_cent_ins R', 'G_and_S_cingul-Ant L', 'G_and_S_cingul-Ant R', 'G_and_S_cingul-Mid-Ant L', 'G_and_S_cingul-Mid-Ant R', 'G_and_S_cingul-Mid-Post L', 'G_and_S_cingul-Mid-Post R', 'G_and_S_frontomargin L', 'G_and_S_frontomargin R', 'G_and_S_occipital_inf L', 'G_and_S_occipital_inf R', 'G_and_S_paracentral L', 'G_and_S_paracentral R', 'G_and_S_subcentral L', 'G_and_S_subcentral R', 'G_and_S_transv_frontopol L', 'G_and_S_transv_frontopol R', 'G_cingul-Post-dorsal L', 'G_cingul-Post-dorsal R', 'G_cingul-Post-ventral L', 'G_cingul-Post-ventral R', 'G_cuneus L', 'G_cuneus R', 'G_front_inf-Opercular L', 'G_front_inf-Opercular R', 'G_front_inf-Orbital L', 'G_front_inf-Orbital R', 'G_front_inf-Triangul L', 'G_front_inf-Triangul R', 'G_front_middle L', 'G_front_middle R', 'G_front_sup L', 'G_front_sup R', 'G_insular_short L', 'G_insular_short R', 'G_oc-temp_lat-fusifor L', 'G_oc-temp_lat-fusifor R', 'G_oc-temp_med-Lingual L', 'G_oc-temp_med-Lingual R', 'G_oc-temp_med-Parahip L', 'G_oc-temp_med-Parahip R', 'G_occipital_middle L', 'G_occipital_middle R', 'G_occipital_sup L', 'G_occipital_sup R', 'G_orbital L', 'G_orbital R', 'G_pariet_inf-Angular L', 'G_pariet_inf-Angular R', 'G_pariet_inf-Supramar L', 'G_pariet_inf-Supramar R', 'G_parietal_sup L', 'G_parietal_sup R', 'G_postcentral L', 'G_postcentral R', 'G_precentral L', 'G_precentral R', 'G_precuneus L', 'G_precuneus R', 'G_rectus L', 'G_rectus R', 'G_subcallosal L', 'G_subcallosal R', 'G_temp_sup-G_T_transv L', 'G_temp_sup-G_T_transv R', 'G_temp_sup-Lateral L', 'G_temp_sup-Lateral R', 'G_temp_sup-Plan_polar L', 'G_temp_sup-Plan_polar R', 'G_temp_sup-Plan_tempo L', 'G_temp_sup-Plan_tempo R', 'G_temporal_inf L', 'G_temporal_inf R', 'G_temporal_middle L', 'G_temporal_middle R', 'Lat_Fis-ant-Horizont L', 'Lat_Fis-ant-Horizont R', 'Lat_Fis-ant-Vertical L', 'Lat_Fis-ant-Vertical R', 'Lat_Fis-post L', 'Lat_Fis-post R', 'Pole_occipital L', 'Pole_occipital R', 'Pole_temporal L', 'Pole_temporal R', 'S_calcarine L', 'S_calcarine R', 'S_central L', 'S_central R', 'S_cingul-Marginalis L', 'S_cingul-Marginalis R', 'S_circular_insula_ant L', 'S_circular_insula_ant R', 'S_circular_insula_inf L', 'S_circular_insula_inf R', 'S_circular_insula_sup L', 'S_circular_insula_sup R', 'S_collat_transv_ant L', 'S_collat_transv_ant R', 'S_collat_transv_post L', 'S_collat_transv_post R', 'S_front_inf L', 'S_front_inf R', 'S_front_middle L', 'S_front_middle R', 'S_front_sup L', 'S_front_sup R', 'S_interm_prim-Jensen L', 'S_interm_prim-Jensen R', 'S_intrapariet_and_P_trans L', 'S_intrapariet_and_P_trans R', 'S_oc-temp_lat L', 'S_oc-temp_lat R', 'S_oc-temp_med_and_Lingual L', 'S_oc-temp_med_and_Lingual R', 'S_oc_middle_and_Lunatus L', 'S_oc_middle_and_Lunatus R', 'S_oc_sup_and_transversal L', 'S_oc_sup_and_transversal R', 'S_occipital_ant L', 'S_occipital_ant R', 'S_orbital-H_Shaped L', 'S_orbital-H_Shaped R', 'S_orbital_lateral L', 'S_orbital_lateral R', 'S_orbital_med-olfact L', 'S_orbital_med-olfact R', 'S_parieto_occipital L', 'S_parieto_occipital R', 'S_pericallosal L', 'S_pericallosal R', 'S_postcentral L', 'S_postcentral R', 'S_precentral-inf-part L', 'S_precentral-inf-part R', 'S_precentral-sup-part L', 'S_precentral-sup-part R', 'S_suborbital L', 'S_suborbital R', 'S_subparietal L', 'S_subparietal R', 'S_temporal_inf L', 'S_temporal_inf R', 'S_temporal_sup L', 'S_temporal_sup R', 'S_temporal_transverse L', 'S_temporal_transverse R'};
58 % Bands for ICC cortex plot
59 BandNames = {'theta', 'alpha', 'beta', 'gamma', 'highgamma'};
60 BandLowerFreqs = [ 4, 8, 13, 30, 50]; % Hz, inclusive
61 BandUpperFreqs = [ 8, 13, 30, 50, 150]; % Hz, exclusive
62
63 %% ===== VERIFY REQUIRED PROTOCOL =====
64 % Check Protocol that it exists
65 iProtocolOmega = bst_get('Protocol', ProtocolNameOmega);
66 if isempty(iProtocolOmega)
67 error(['Unknown protocol: ' ProtocolNameOmega]);
68 end
69 % Select input protocol
70 gui_brainstorm('SetCurrentProtocol', iProtocolOmega);
71
72 % Verify the Subjects
73 ProtocolSubjects = bst_get('ProtocolSubjects');
74 ProtocolSubjectNames = {ProtocolSubjects.Subject.Name};
75 if ~all(ismember(SubjectNames, ProtocolSubjectNames))
76 error(['All requested subjects must be present in the ' ProtocolNameOmega ' protocol.']);
77 end
78
79
80 %% ===== FIND FILES =====
81 bst_report('Start');
82
83 % Get raw and source files for each Subject
84 sRawDataFiles = [];
85 sSourcesFiles = [];
86
87 for iSubject = 1 : nSubjects
88 % Process: Select data files in: sub-000*/*
89 sFiles = bst_process('CallProcess', 'process_select_files_data', [], [], ...
90 'subjectname', SubjectNames{iSubject}, ...
91 'condition', '', ...
92 'tag', '', ...
93 'includebad', 0, ...
94 'includeintra', 0, ...
95 'includecommon', 0);
96 sRawDataFiles = [sRawDataFiles, sFiles];
97
98 % Process: Select results files in: sub-000*/*
99 sFiles = bst_process('CallProcess', 'process_select_files_results', [], [], ...
100 'subjectname', SubjectNames{iSubject}, ...
101 'condition', '', ...
102 'tag', '', ...
103 'includebad', 0, ...
104 'includeintra', 0, ...
105 'includecommon', 0);
106 sSourcesFiles = [sSourcesFiles, sFiles];
107 end
108
109
110 %% Compute PSD for all ROIs of an atlas
111 % For each subject, their recordings are split in two parts (data segments)
112 % these two data segments will be used to create PSDs
113 % These two PSDs are the features which will define the brain-fingerprint
114
115 nSegments = 2; % Training and Validation
116 timeIni = zeros(nSubjects, nSegments);
117 timeFin = zeros(nSubjects, nSegments);
118 sPsdFiles = repmat(db_template('processfile'), nSubjects, nSegments);
119 for iSubject = 1 : nSubjects
120 sData = load(file_fullpath(sRawDataFiles(iSubject).FileName), 'Time');
121 halfTime = diff(sData.Time) / 2;
122 % First segment
123 timeIni(iSubject, 1) = sData.Time(1) + 30;
124 timeFin(iSubject, 1) = halfTime - 30;
125 % Second segment
126 timeIni(iSubject, 2) = halfTime + 30;
127 timeFin(iSubject, 2) = sData.Time(2) - 30;
128 % Compute PSD
129 for iSegment = 1 : size(timeIni, 2)
130 % Process: Power spectrum density (Welch)
131 sPsdFiles(iSubject, iSegment) = bst_process('CallProcess', 'process_psd', sSourcesFiles(iSubject).FileName, [], ...
132 'timewindow', [timeIni(iSubject, iSegment), timeFin(iSubject, iSegment)], ...
133 'win_length', 2, ...
134 'win_overlap', 50, ...
135 'units', 'physical', ... % Physical: U2/Hz
136 'clusters', {AtlasName, AtlasScouts}, ...
137 'scoutfunc', 1, ... % Mean
138 'win_std', 0, ...
139 'edit', struct(...
140 'Comment', 'Scouts,Power', ...
141 'TimeBands', [], ...
142 'Freqs', [], ...
143 'ClusterFuncTime', 'before', ...
144 'Measure', 'power', ...
145 'Output', 'all', ...
146 'SaveKernel', 0));
147 end
148 end
149
150
151 %% ===== VECTORIZE PSD DATA FOR FINGERPRINTING =====
152 % Find size of requested PSD data
153 sPsdMat = in_bst_timefreq(sPsdFiles(1,1).FileName, 0, 'RowNames', 'Freqs');
154 Freqs = sPsdMat.Freqs;
155 ixLowerFreq = find(Freqs >= LowerFreq, 1, 'first');
156 ixUpperFreq = find(Freqs < UpperFreq, 1, 'last');
157 Freqs = sPsdMat.Freqs(ixLowerFreq:ixUpperFreq);
158 nFreqs = (ixUpperFreq - ixLowerFreq + 1);
159 ScoutNames = sPsdMat.RowNames;
160 nScouts = length(ScoutNames);
161
162 % Subject vectors
163 trainingVectors = zeros(iSubject, nScouts * nFreqs);
164 validationVectors = zeros(iSubject, nScouts * nFreqs);
165
166 % Vectorize Scout PSDs
167 for iSubject = 1 : nSubjects
168 for iSegment = 1 : nSegments
169 sPsdMat = in_bst_timefreq(sPsdFiles(iSubject, iSegment).FileName, 0, 'TF');
170 if iSegment == 1
171 trainingVectors(iSubject,:) = reshape(squeeze(sPsdMat.TF(:, :, ixLowerFreq:ixUpperFreq)), 1, []);
172 elseif iSegment == 2
173 validationVectors(iSubject,:) = reshape(squeeze(sPsdMat.TF(:, :, ixLowerFreq:ixUpperFreq)), 1, []);
174 end
175 end
176 end
177
178
179 %% ==== SIMILARITY AND DIFFERENTIABILITY =====
180 % Subject similarity matrix
181 % It is generally symmetric although it does not necessarily have to be!
182 SubjectCorrMatrix= corr(trainingVectors', validationVectors');
183 % Differentiability
184 diff_1= (diag(SubjectCorrMatrix)-mean(SubjectCorrMatrix,1) )/ std(SubjectCorrMatrix,0,1); % along columns
185 diff_2= (diag(SubjectCorrMatrix)-mean(SubjectCorrMatrix,2)')/ std(SubjectCorrMatrix,0,2)'; % along rows
186 % Differentiability across rows and columns are generally strongly correlated
187 % Take the mean for a summary statistic per participant
188 Differentiability = (diff_1 + diff_2)/2; % Mean differentiability derived from rows and columns
189
190 % IntrClass Correlations (ICC)
191 zTraining = (trainingVectors - mean(trainingVectors, 2)) ./ std(trainingVectors, 0, 2);
192 zValidation= (validationVectors - mean(validationVectors, 2)) ./ std(validationVectors, 0, 2);
193 df_b = nSubjects-1; % degrees of freedom for s2
194 df_w = nSubjects*(nSegments-1); % degrees of freedom for r
195 nFeatures = nScouts * nFreqs;
196 icc = zeros(1, nFeatures);
197 for iFeature = 1 : nFeatures
198 x = [zTraining(:, iFeature), zValidation(:, iFeature)];
199 x_w_mean = mean(x, 2);
200 x_g_mean = mean(x(:));
201 ss_t = sum((x - x_g_mean).^2, 'all');
202 ss_w = sum((x - x_w_mean).^2, 'all');
203 ss_b = ss_t - ss_w;
204 ms_b = ss_b / df_b;
205 ms_w = ss_w / df_w;
206 icc(iFeature) = (ms_b - ms_w) ./ (ms_b + ((nSegments-1).*ms_w));
207 end
208 iccFreqsScouts = reshape(icc, [nFreqs, nScouts]);
209 % Compute Scout ICC for frequency bands
210 nBands = length(BandNames);
211 iccBands = zeros(nScouts, nBands);
212 for iBand = 1 : nBands
213 ixLowerFreq = find(Freqs >= BandLowerFreqs(iBand), 1, 'first');
214 ixUpperFreq = find(Freqs < BandUpperFreqs(iBand), 1, 'last');
215 iccBands(: ,iBand) = mean(iccFreqsScouts(ixLowerFreq:ixUpperFreq, :), 1)';
216 end
217
218 %% ===== SAVE OUTCOME IN BRAINSTORM DATABASE =====
219 % Retrieve 'Group_analysis' subject
220 sNormSubj = bst_get('NormalizedSubject');
221 % Study to save files
222 [sOutputStudy, iOutputStudy] = bst_get('StudyWithCondition', bst_fullfile(sNormSubj.Name, bst_get('DirAnalysisIntra')));
223
224 % Similarity matrix
225 sSimilarityMat = db_template('timefreqmat');
226 % Reshape: [nA x nB x nTime x nFreq] => [nA*nB x nTime x nFreq]
227 sSimilarityMat.TF = reshape(SubjectCorrMatrix, [], 1, 1);
228 sSimilarityMat.Comment = 'Similarity matrix';
229 sSimilarityMat.DataType = 'matrix';
230 sSimilarityMat.Time = [0, 1];
231 sSimilarityMat.RefRowNames = cellfun(@(x) ['Train ', x], SubjectNames, 'UniformOutput', false);
232 sSimilarityMat.RowNames = cellfun(@(x) ['Validation ', x], SubjectNames, 'UniformOutput', false);
233 % Output filename
234 SimilarityFile = bst_process('GetNewFilename', bst_fileparts(sOutputStudy.FileName), 'timefreq_connectn_corr');
235 % Save file
236 bst_save(SimilarityFile, sSimilarityMat, 'v6');
237 % Add file to database structure
238 db_add_data(iOutputStudy, SimilarityFile, sSimilarityMat);
239
240 % Differentiability matrix
241 sDiffMat = db_template('matrixmat');
242 sDiffMat.Value = Differentiability;
243 sDiffMat.Comment = 'Differentiability';
244 sDiffMat.Time = [0, 1];
245 sDiffMat.Description = SubjectNames;
246 % Output filename
247 DiffFile = bst_process('GetNewFilename', bst_fileparts(sOutputStudy.FileName), 'matrix');
248 % Save file
249 bst_save(DiffFile, sDiffMat, 'v6');
250 % Add file to database structure
251 db_add_data(iOutputStudy, DiffFile, sDiffMat);
252
253 % Save band ICC values (scout level)
254 for iBand = 1 : nBands
255 sIccBandMat = db_template('matrixmat');
256 sIccBandMat.Value = iccBands(:, iBand);
257 sIccBandMat.Comment = ['ICC_' BandNames{iBand}];
258 sIccBandMat.Time = [0, 1];
259 sIccBandMat.Description = ScoutNames;
260 % Output filename
261 IccBandFile = bst_process('GetNewFilename', bst_fileparts(sOutputStudy.FileName), 'matrix');
262 % Save file
263 bst_save(IccBandFile, sIccBandMat, 'v6');
264 % Add file to database structure
265 db_add_data(iOutputStudy, IccBandFile, sIccBandMat);
266 end
267
268 % Cortex surface display for band ICC values
269 % Verify destination surface
270 sSubjectGroup = bst_get('Subject', sOutputStudy.BrainStormSubject);
271 sSurfFile = sSubjectGroup.Surface(sSubjectGroup.iCortex);
272 sSurfMat = in_tess_bst(sSurfFile.FileName);
273 iAtlas = find(ismember({sSurfMat.Atlas.Name}, AtlasName));
274 if isempty(iAtlas)
275 error('Default surface in "%s" does not contain "%s" atlas', bst_get('NormalizedSubjectName'), AtlasName);
276 end
277 sAtlas = sSurfMat.Atlas(iAtlas);
278 if ~all(ismember({sAtlas.Scouts.Label}, ScoutNames))
279 error('Atlas "%s" in "%s" does not contain the same scouts as ICC', AtlasName, bst_get('NormalizedSubjectName'));
280 end
281 % Create one full results file per band ICC values
282 IccBandFiles = {};
283 for iBand = 1 : nBands
284 sIccBandMat = db_template('resultsmat');
285 sIccBandMat.SurfaceFile = sSurfFile.FileName;
286 sIccBandMat.ImageGridAmp = nan(size(sSurfMat.Vertices, 1), 1);
287 sIccBandMat.Comment = ['ICC_' BandNames{iBand}];
288 sIccBandMat.Time = [0, 1];
289 % Assign ICC values to vertices in Scout
290 for ix = 1 : nScouts
291 iScout = find(ismember({sAtlas.Scouts.Label}, ScoutNames(ix)));
292 sIccBandMat.ImageGridAmp(sAtlas.Scouts(iScout).Vertices) = deal(iccBands(ix, iBand));
293 end
294 % Output filename
295 IccBandFile = bst_process('GetNewFilename', bst_fileparts(sOutputStudy.FileName), 'results');
296 IccBandFiles = [IccBandFiles, IccBandFile];
297 % Save file
298 bst_save(IccBandFile, sIccBandMat, 'v6');
299 % Add file to database structure
300 db_add_data(iOutputStudy, IccBandFile, sIccBandMat);
301 end
302
303 % Reload database
304 db_reload_studies(iOutputStudy)
305
306
307 %% ===== SNAPSHOTS =====
308 % Process: Snapshot: Similarity matrix
309 bst_process('CallProcess', 'process_snapshot', SimilarityFile, [], ...
310 'type', 'connectimage', ... % Connectivity matrix
311 'Comment', 'Similarity matrix');
312
313 % Process: Snapshot: Recordings time series
314 bst_process('CallProcess', 'process_snapshot', DiffFile, [], ...
315 'type', 'data', ... % Recordings time series
316 'time', 0, ...
317 'Comment', 'Differentiability');
318
319 for iBand = 1 : nBands
320 % Process: Snapshot: Sources (one time)
321 bst_process('CallProcess', 'process_snapshot', IccBandFiles{iBand}, [], ...
322 'type', 'sources', ... % Sources (one time)
323 'orient', 1, ... % left
324 'time', 0, ...
325 'contact_time', [0, 0.1], ...
326 'contact_nimage', 12, ...
327 'threshold', 0, ...
328 'surfsmooth', 30, ...
329 'mni', [0, 0, 0], ...
330 'Comment', ['ICC_' BandNames{iBand}]);
331 end
332
333 % Save report
334 ReportFile = bst_report('Save', []);
335 if ~isempty(reports_dir) && ~isempty(ReportFile)
336 bst_report('Export', ReportFile, reports_dir);
337 else
338 bst_report('Open', ReportFile);
339 end



