Tutorial 2: Import the subject anatomy
Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet
Contents
Download
The dataset we will use for the introduction tutorials is available online.
Go to the Download page of this website, and download the file: sample_introduction.zip
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder).
- This is really important that you always keep your original data files in a separate folder: the program folder can be deleted when updating the software, and the contents of the database folder is supposed to be manipulated only by the program itself.
Create a new subject
The protocol is currently empty. You need to add a new subject before you can start importing data.
- Switch to the anatomy view (first button just above the database explorer).
Right-click on the top folder TutorialIntroduction > New subject.
Alternatively: Use the menu File > New subject.
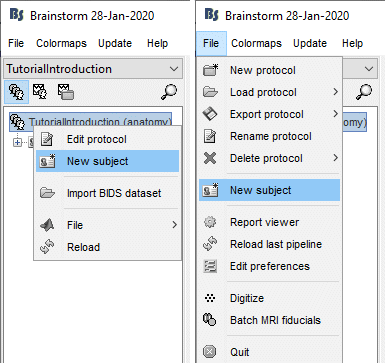
The window that opens lets you edit the subject name and settings. It offers again the same options for the default anatomy and channel file: you can redefine for one subject the default values set at the protocol level if you need to.
- Keep all the default settings and click on [Save].
Right-click doesn't work
If the right-click doesn't work anywhere in the Brainstorm interface and you cannot get to see the popup menus in the database explorer, try to connect a standard external mouse with two buttons. Some Apple pointing devices do not interact very well with Java/Matlab.
Alternatively, try to change the configuration of your trackpad in the system preferences.
Import the anatomy
For estimating the brain sources of the MEG/EEG signals, the anatomy of the subject must include at least three files: an MRI volume, the envelope of the cortex and the head surface of the head.
Brainstorm cannot extract the cortex envelope from the MRI, you have to run this operation with an external program of your choice. The results of the MRI segmentation obtained with the following programs can be automatically imported: FreeSurfer, BrainSuite, BainVISA and CIVET.
The anatomical information of this study was acquired with a 1.5T MRI scanner, the subject had a marker placed on the left cheek. The MRI volume was processed with FreeSurfer 5.3, the result of this automatic segmentation process is available in the downloaded folder sample_introduction/anatomy.
- Make sure that you are still in the anatomy view for your protocol.
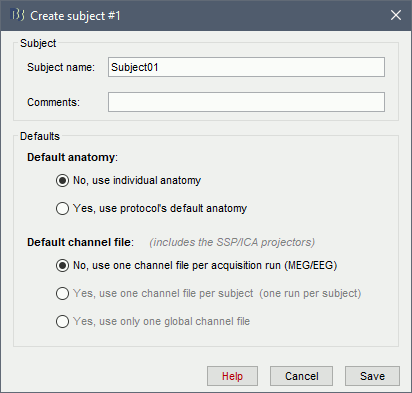
Right-click on the subject folder > Import anatomy folder:
Set the file format: FreeSurfer folder
Select the folder: sample_introduction/anatomy
- Click on [Open]
Number of vertices of the cortex surface: 15000 (default value)
This option defines the number of points that will be used to represent the cortex envelope. It will also be the number of electric dipoles we will use to model the activity of the brain. This default value of 15000 was chosen empirically a good balance between the spatial accuracy of the models and the computation speed. More details later in the tutorials.
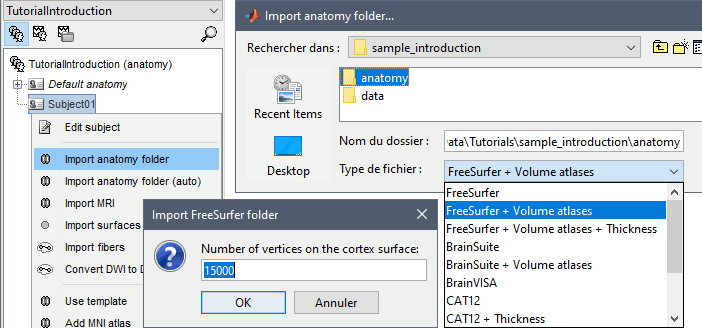
The MRI viewer is displayed, together with a message box that tells you what to do. Follow these instructions. The MRI views should be correct (axial/coronal/sagittal), you just need to make sure that the marker on the cheek is really on the left of the MRI. Then you can proceed with the fiducial selection.
Using the MRI Viewer
To help define these fiducial points, let's start with a brief description of the MRI Viewer:
Navigate in the volume:
- Click anywhere on the MRI slices to move the cursor.
- Use the sliders below the views.
- Use the mouse wheel to scroll through slices (after clicking on the view to select it).
On a MacBook pad, use the two finger-move up/down to scroll.
Zoom: Use the magnifying glass buttons at the bottom of the figure.
Image contrast: Click and hold the right mouse button on one image, then move up and down.
Select a point: Place the cursor at the spot you want and click on the corresponding [Set] button.
Fiducial points
Brainstorm uses a few reference points defined in the MRI to align the different files:
- Three to define the Subject Coordinate System (SCS):
- Nasion (NAS), Left ear (LPA), Right ear (RPA)
- This is used to register the MEG/EEG sensors on the MRI.
- Three to define the Normalized coordinate system (NCS):
- Anterior commissure (AC), Posterior commissure (PC) and any interhemispheric point (IH)
- This is used to align the individual subject's anatomy on the anatomical templates.
- When using one of the templates that has been registered to the MNI stereotaxic space (Colin27 or ICBM152), these coordinates correspond to the standard MNI coordinates.
For instructions on finding these points, read the following page: CoordinateSystems.
Nasion (NAS)
In this study, we used the real nasion position instead of the CTF coil position.
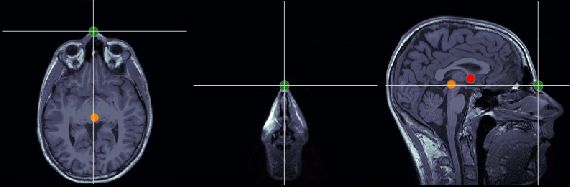
MRI coordinates: 127, 213, 139
Left ear (LPA)
In this study, we used the connection points between the tragus and the helix (red dot on the CoordinateSystems page) instead of the CTF coil position or the left and right preauricular points.
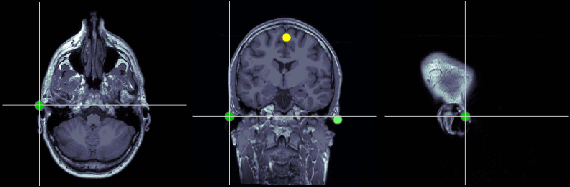
MRI coordinates: 52, 113, 96
Right ear (RPA)
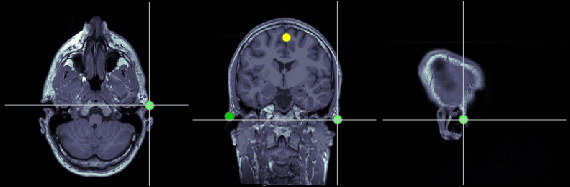
MRI coordinates: 202, 113, 91
Anterior commissure (AC)
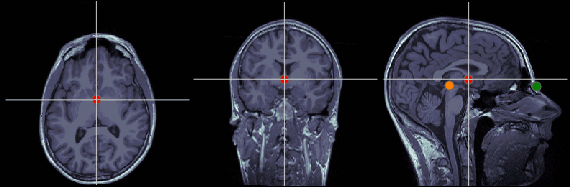
MRI coordinates: 127, 119, 149
Posterior commissure (PC)
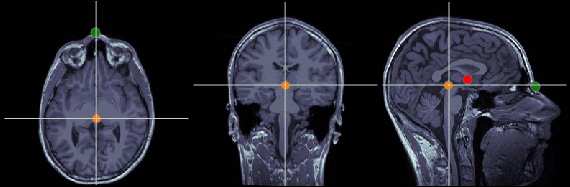
MRI coordinates: 128, 93, 141
Inter-hemispheric point (IH)
This point can be anywhere in the mid-sagittal plane, these coordinates are just an example.
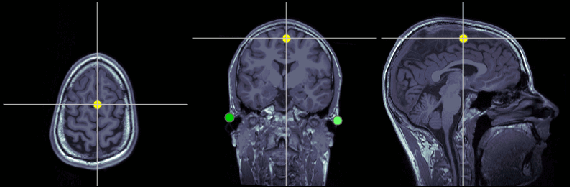
MRI coordinates: 131, 114, 206
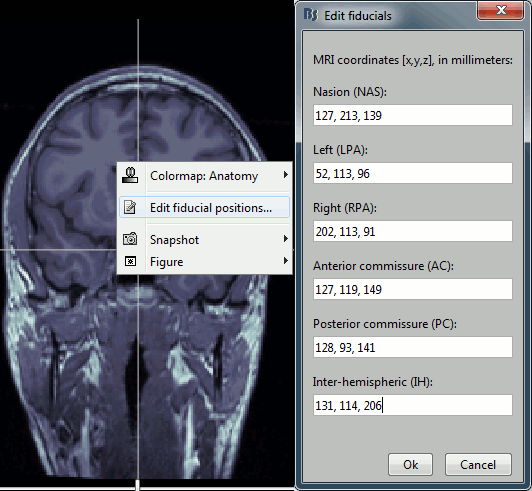
Type the coordinates
If you have the coordinates of the fiducials already written somewhere, you can type or copy-paste them instead of the pointing at them in with the cursor. Right-click on the figure > Edit fiducials positions.
Validation
- Once you are done with the fiducial selection, click on [Save].
The automatic import of the FreeSurfer folder resumes. At the end you get many new files in the database and a 3D view of the cortex and scalp surface. Here again you can note that the marker is visible on the left cheek, as expected.
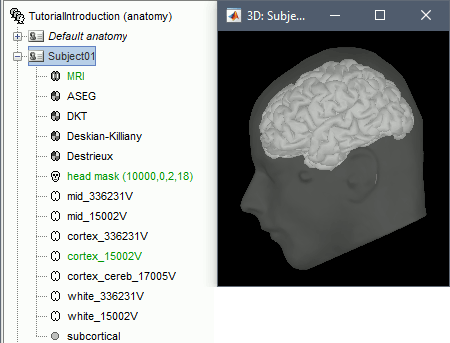
- The next tutorial will describe these files and explore the various visualization options.
Close all figures and clear memory: Use this button in the toolbar of the Brainstorm window to close all the open figures at once and to empty the memory from all the temporary data that the program keeps in memory for faster display.
Graphic bugs
If you do not see the cortex surface through the head surface, or if you observe any other issue with the 3D display, there might be an issue with the OpenGL drivers. You may try the following options:
- Update the drivers for your graphics card.
- Upgrade your version of Matlab.
Run the compiled version of Brainstorm (see Installation).
Turn off the OpenGL hardware acceleration: Menu File > Edit preferences > Software or Disabled.
- Send a bug report to the Mathworks.
MNI transformation
For comparing results with the literature or with other imaging modalities, the normalized MNI coordinate system is often used. To be able to get "MNI coordinates" for individual brains, an extra step of normalization is required. The method we use in Brainstorm is based on an affine co-registration with the MNI ICBM152 template from the SPM software, described in the following article: Ashburner J, Friston KJ, Unified segmentation, NeuroImage 2005.
To compute the linear transformation matrix between the individual MRI and the ICBM152 template, you have two available options, use the one of your choice:
- In the MRI Viewer: Click on the link "Click here to compute MNI transformation".
In the database explorer: Right-click on the MRI > Compute MNI transformation.
Note that this transformation does not modify the anatomy, it just saves an affine transformation that enables the conversion between Brainstorm coordinates and MNI coordinates. After computing this transformation, you have access to one new line of information in the MRI Viewer.
This operation also sets automatically some anatomical points (AC, PC, IH). After the computation, make sure they are correctly positioned. You can also run this computation while importing the anatomy, when the MRI viewer is displayed for the first time, this will save you the trouble of marking the AC/PC/IH points manually.
Alternatives
If you do not have access to an individual MR scan of the subject, or if its quality is too low to be processed with FreeSurfer, you have other options:
If you do not have any individual anatomical data: Use the default anatomy
If you have a digitized head shape of the subject: Warp the default anatomy
