MEG current phantom (Elekta-Neuromag)
Authors: Ken Taylor, John Mosher
This tutorial explains how to import and process Elekta-Neuromag current phantom recordings. We decided to release this example for testing and cross-validation purposes. With these datasets, we can evaluate the equivalence of various forward models and dipole fitting methods in the case of simple recordings with single dipoles. The recordings are available in two file formats (native and FIF) to cross-validate the file readers available in Brainstorm and MNE. A similar page exists for the CTF phantom.
Contents
License
This tutorial dataset remains a property of its authors: Ken Taylor, John Mosher (Epilepsy Center, Cleveland Clinic Neurological Institute, Cleveland, OH USA).
If you reference this dataset in your publications, please acknowledge them and cite Brainstorm as indicated on the website. For questions, please contact us through the forum.
The phantom
A current phantom is provided with the Elekta Neuromag for checking the system performance. It contains 32 artificial dipoles and four fixed head-position indicator coils. The phantom is based on the mathematical fact that an equilateral triangular line current produces equivalent magnetic field distribution to that of a tangential current dipole in a spherical conductor, provided that the vertex of the triangle and the origin of the conducting sphere coincide. For a detailed description of how the phantom works, see here.
The phantom dipoles are energized using an internal signal generator which also feeds the HPI coils. An external multiplexer box is used to connect the signal to the individual dipoles. Only one dipole can be activated at a time. The location of the dipole is recorded relative to the center of the sphere (0,0,0)m, where X is positive toward the nasion, Y is positive toward the left ear and Z is positive toward the top of the head (see the CoordinateSystems tutorial for more details).
Use of the phantom is shown below. Note that the uncovered version is the phantom that came with the Neuromag-122, which explicitly shows the wiring. The covered version uses the same principle but somewhat different dipole locations. Further details are available in Section 7.2 of the User's Manual.
References
R.J. Ilmoniemi, M.S. Hämäläinen, and J. Knuutila, The Forward and Inverse Problems in the Spherical Model. In: Biomagnetism: Applications and Theory, eds. H. Weinberg, G. Stroink, T. Katil, Pergamon Press, 1985.
Elekta Neuromag System Hardware User's Manual, Revision G, September 2005.
Description of the experiment
[To do]
Download and installation
Requirements: You have followed the introduction tutorials and Brainstorm is installed.
Go to the Download page of this website, and download the file: sample_phantom_elekta.zip
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder)
- Start Brainstorm (Matlab scripts or stand-alone version)
Select the menu File > Create new protocol. Name it TutorialPhantomElekta and select the options:
No, use individual anatomy,
No, use one channel file per acquisition run (MEG).
Generate anatomy
In the Matlab command window: type "generate_phantom_elekta".
This creates a new subject Kojak_Sphere (so named after the 70's TV show) and generates the "anatomy" for this device: one volume and a few surfaces representing the geometry of the phantom.
Access the recordings
We can now review one of the raw kojak data sets. These have been generated by sequentially activating each of the 32 phantom dipoles in a single raw file.
Switch to the functional data view by clicking the second button under the protocol name.
Right-click on the subject folder > Review raw file.
Select the 200nA source.
Click Event channel, select STI201, and hit OK.
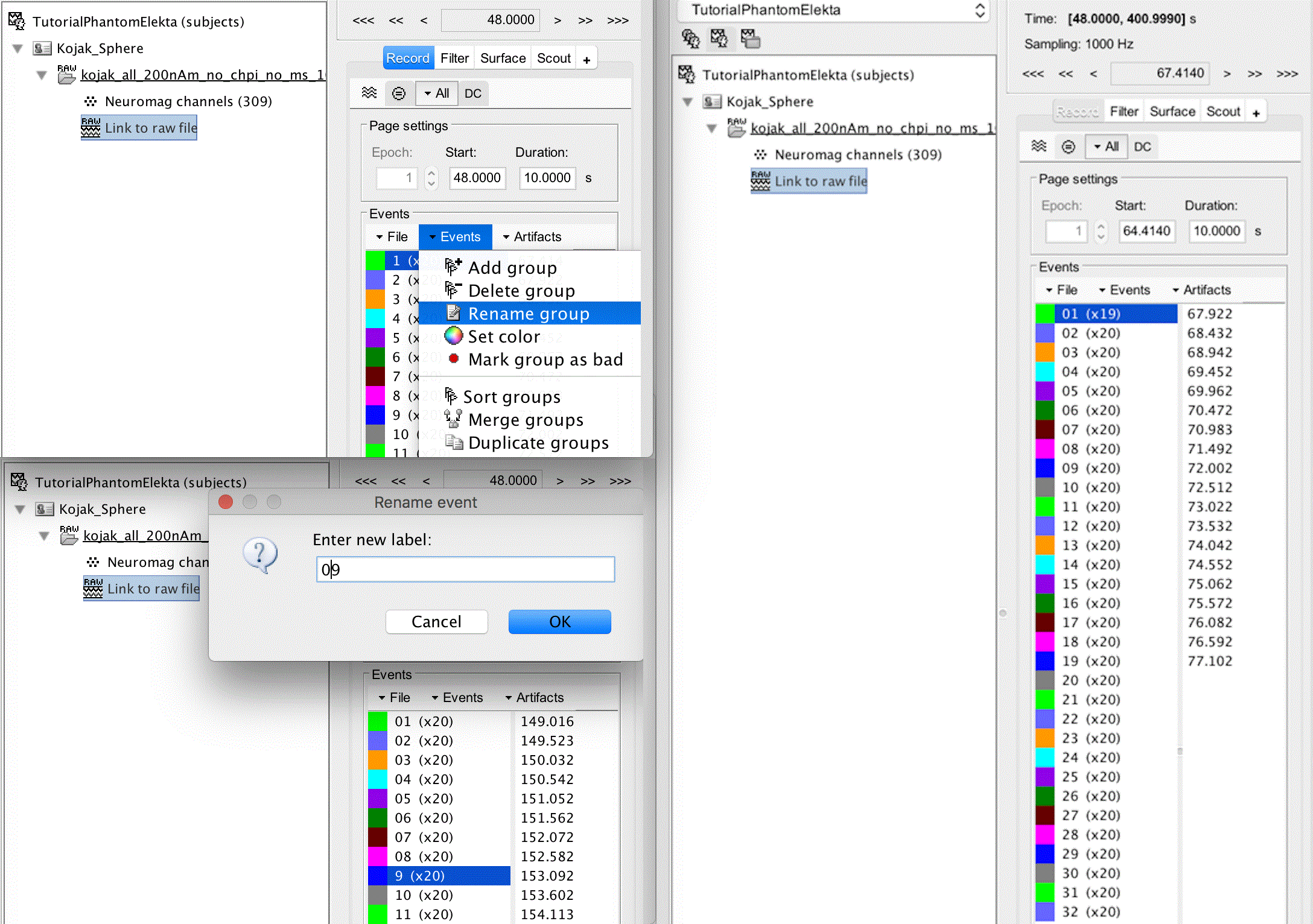
Close the figure and double click the link to raw file to open a list of the events. At this point, you may wish to rename your groups so that the ordering remains convenient. To do this, click group 1, click Events > Rename group, and rename it to 01. Repeat this for 2 - 9.
This step, and others like it, can also be performed by importing the data into MATLAB, running the following code below, and then exporting the data back into Brainstorm:
% to change the trigger numbers to appear sequentially
for i = 1:32,
STR = str2num(link_filename.F.events(i).label);
link_filename.F.events(i).label = sprintf('%02.0f',STR);
endSome other triggers are generated by the neuromag system which are unnecessary so we will delete them:
Trim away the singleton events by clicking on 256 (x1), shift clicking on 7936 (x1) and pressing the delete key.
There is also a switching transient in the phantom generator which should be removed:
Remove the first event from dipole 01 by clicking on it, clicking on the first time instant, and pressing the delete key.
Events should now appear as per the image below on the right:
Note: Make sure to save the modifications that you make before proceeding. If you hit the grey X to close all figures and clear memory, you will be prompted to save modifications, and you should click Yes.
Importing the dipole events
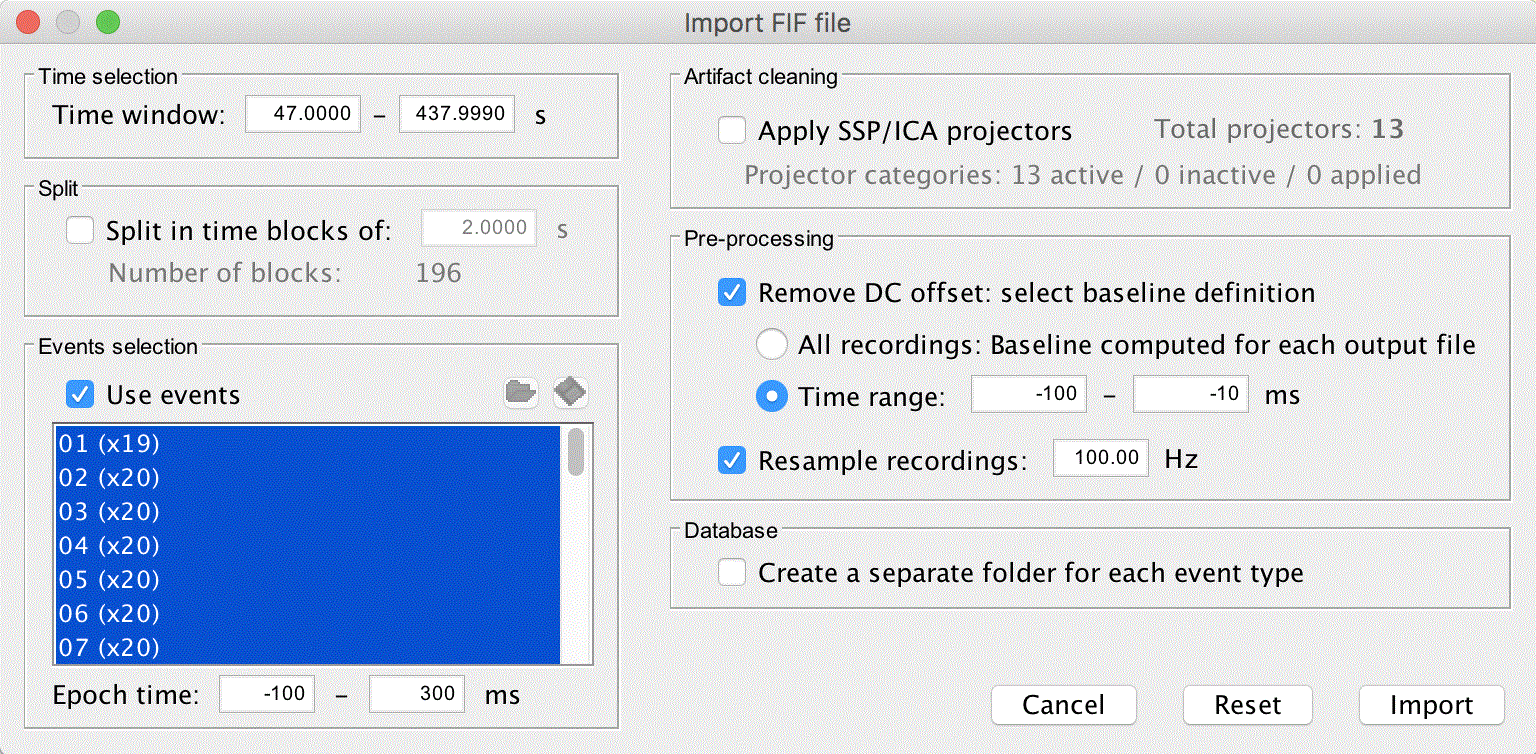
Right click on Link to raw file and click Import in database
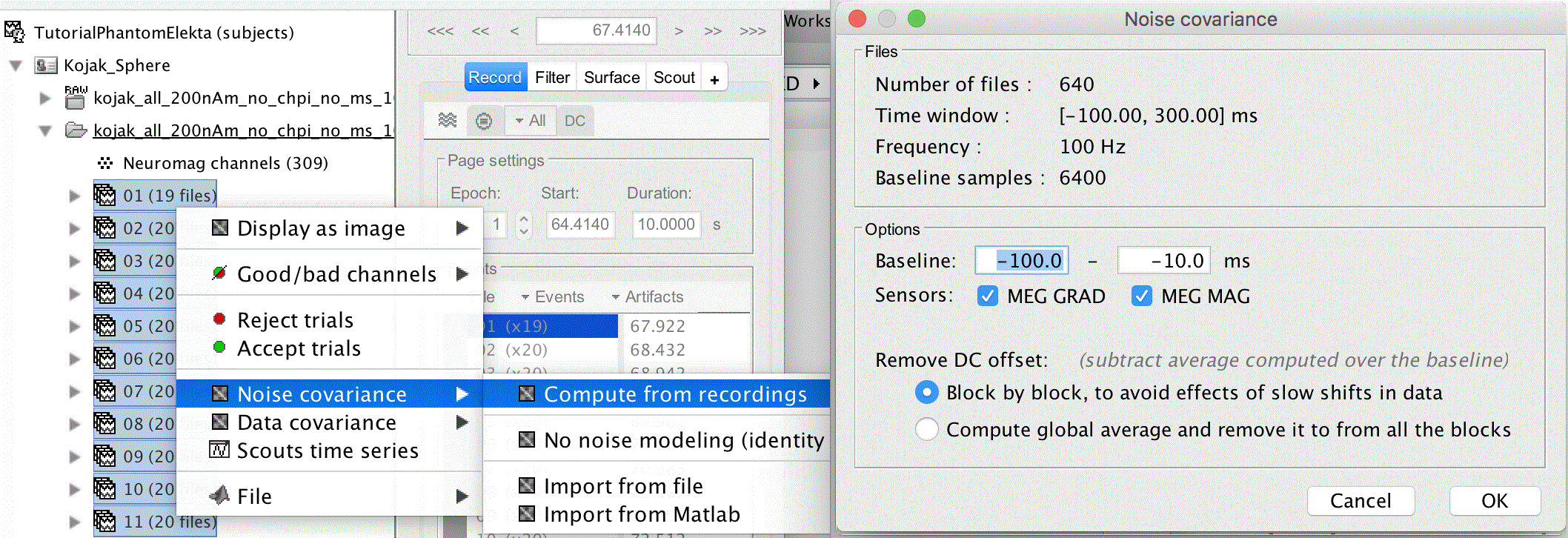
Uncheck Apply SSP/ICA projectors (we will use noise covariance to stabilize the data)
Check Remove DC offset, set the time range from -100 to -10ms
Resample the recordings to 100Hz (change from 1000Hz)
Do not create separate folders
Click Import
Noise covariance
With the epochs loaded, we can now calculate the noise covariance from the prestims. To do this, select all 32 dipoles (click the first then shift click the last), then right click, Noise covariance > Compute from recordings. Uncheck MEG MAG and hit OK.
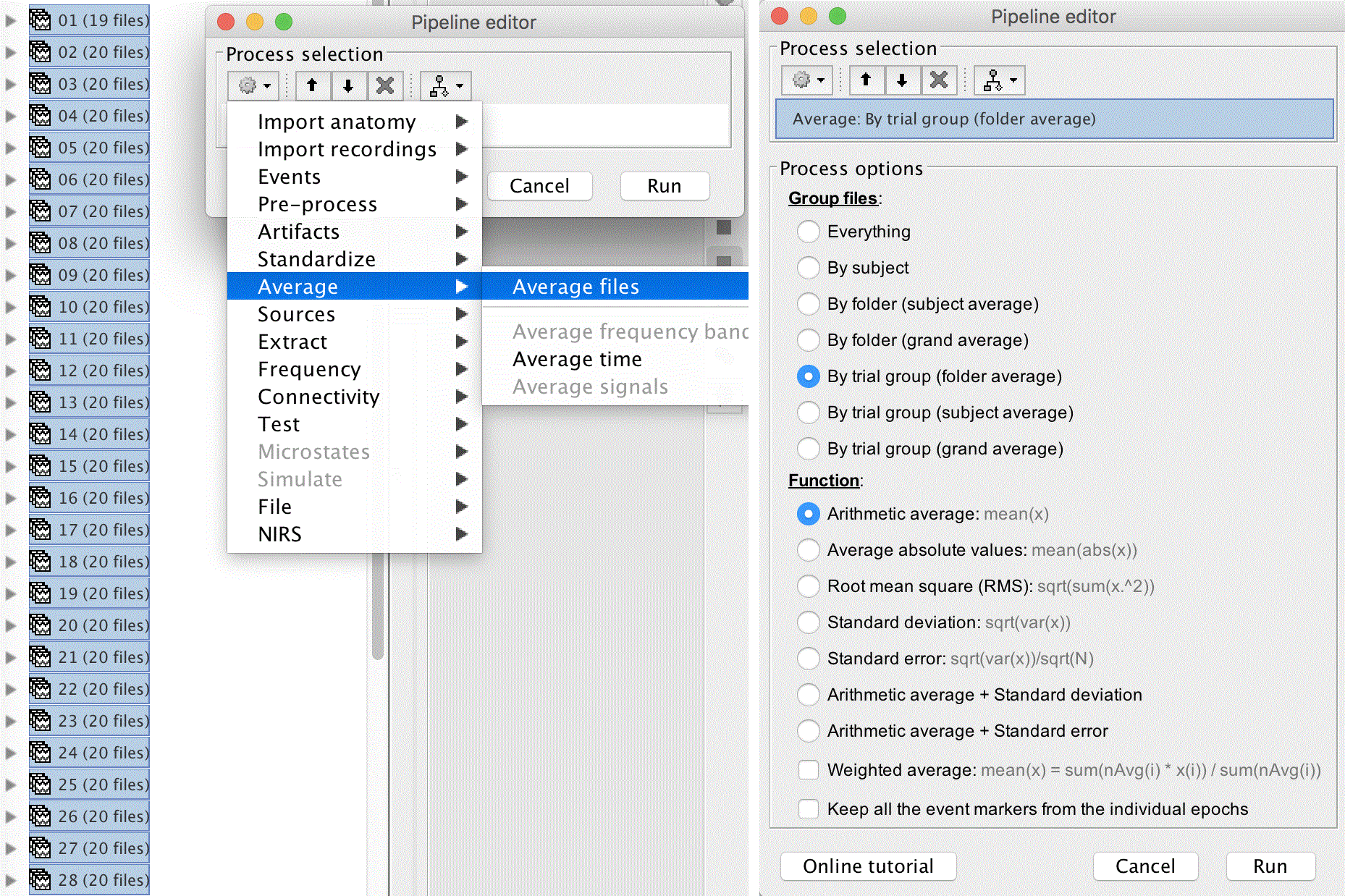
Average all of the epochs by selecting all 32 again, dragging them to the process box, clicking RUN. Next click the cog, and select Average > Average files. Group the files By trial group (folder average), and select Arithmetic average: mean(x), and click "Run".
Head modelling
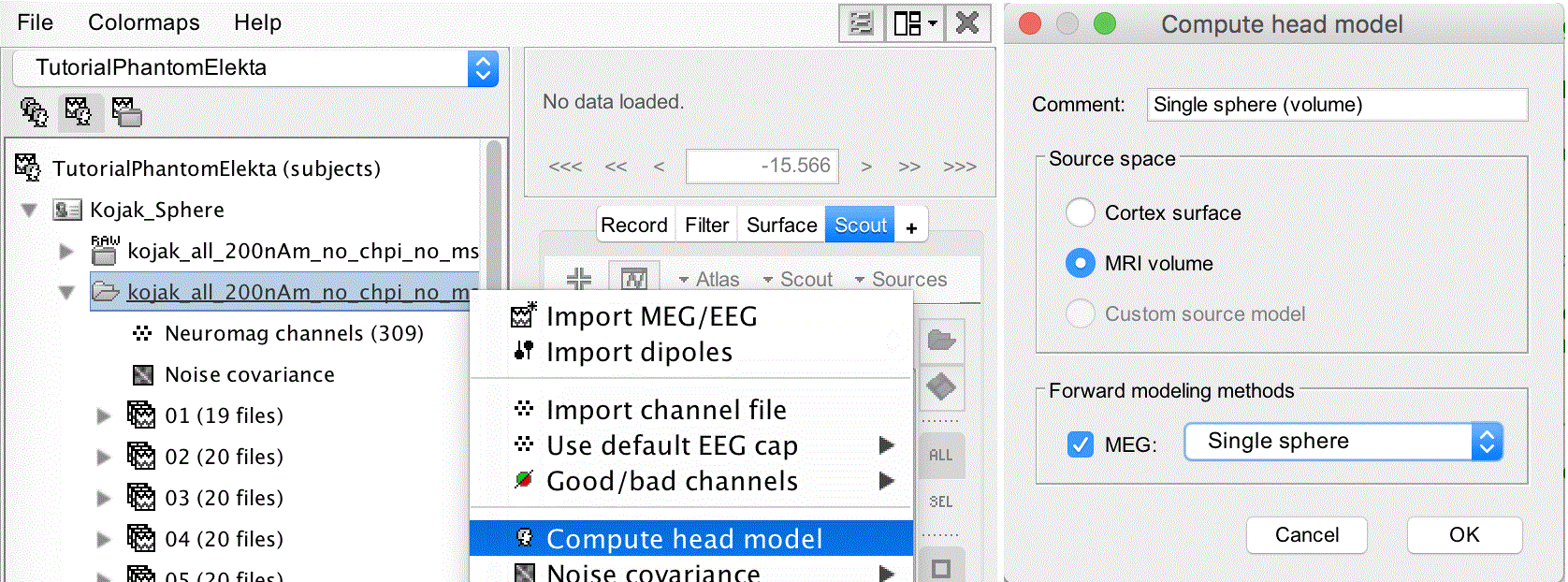
To compute the head model, right click on the kojak_all_200nA file and select Compute head model.
Source space: select MRI volume
Forward modeling methods: Single sphere
A figure opens showing a sphere fit to the phantom, however we can see that the location is a little off. In this case we alread know where the center of the sphere should be, so we adjust it manually:
Click the Edit sphere properties button in the top toolbar.
Change the center location to [ 0 0 0 ] and hit OK.
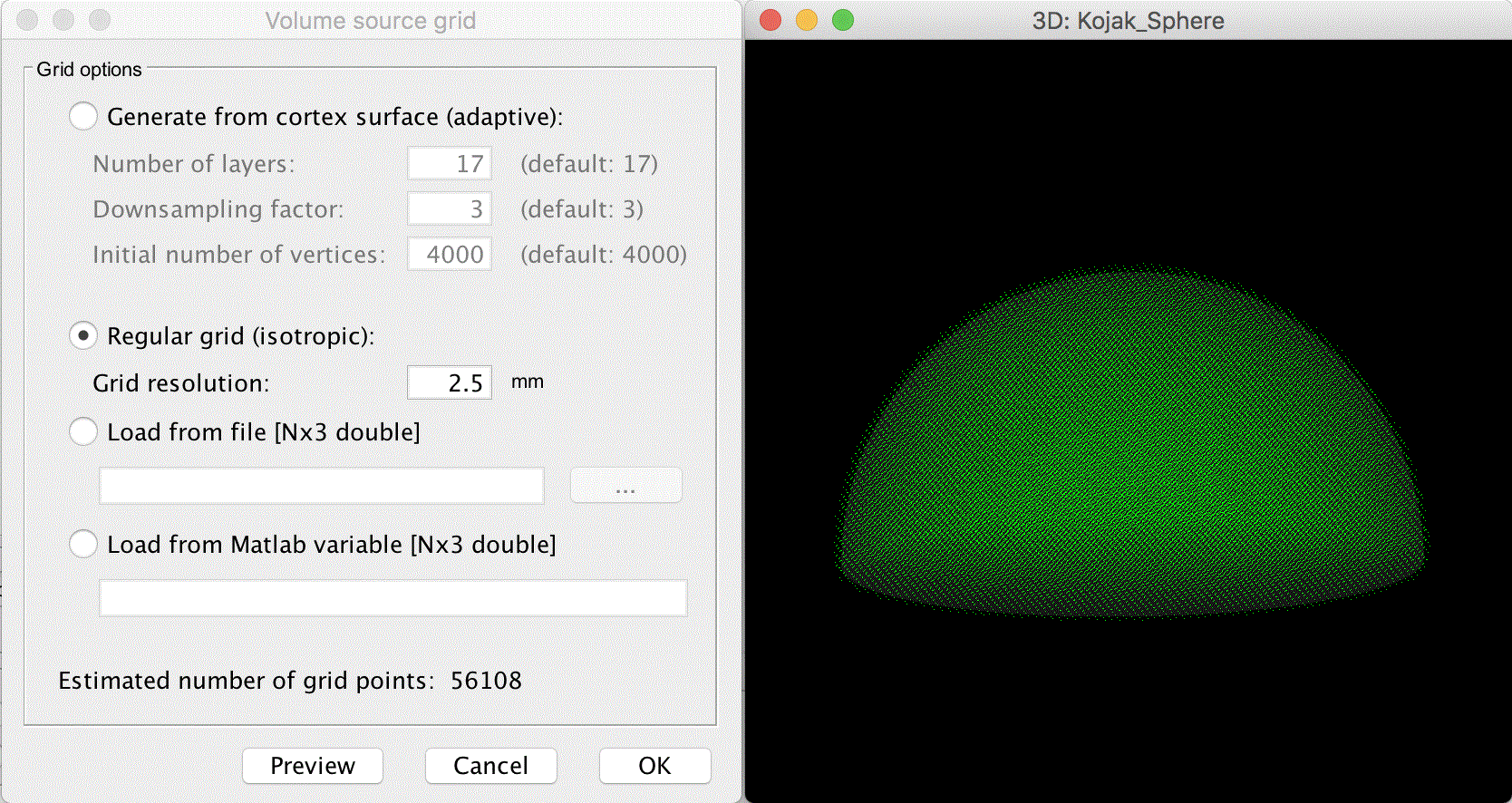
The center of the sphere shifts to the correct position. Click the OK button next to the edit sphere properties button to perform the MRI/surface interpolation. A window appears allowing adjustment of the volume source grid.
Select Regular grid (isotropic) with Grid resolution 2.5mm
This results in an around 56K grid point, which is sufficient.
Dipole source estimation
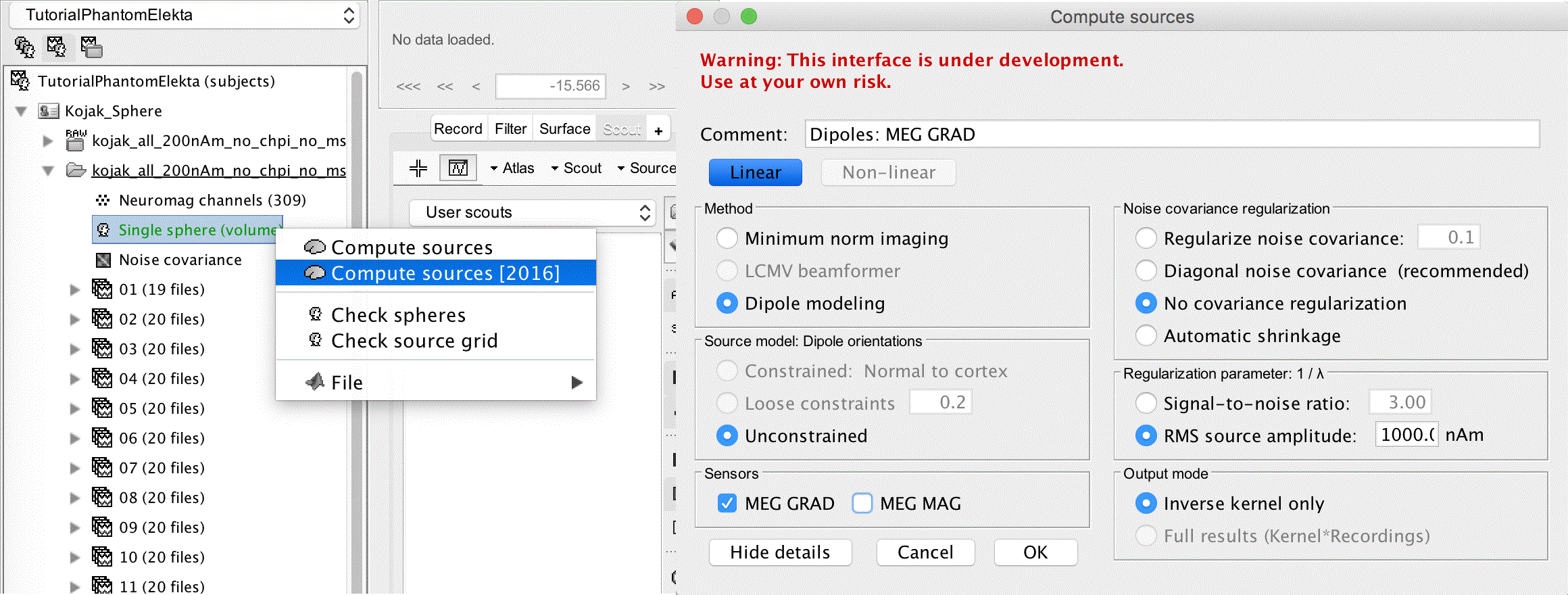
Right click the Single sphere (volume) and select Compute sources [2016]
Method: Dipole modeling
Source model: Dipole orientations Unconstrained
For the sensor selection you can choose to use either the gradiometers or the magnetometers, in this case we select both.
Click Show details if the noise covariance regularization is hidden.
Select Regularize noise covariance: 0.1
Ignore the other settings, and click OK
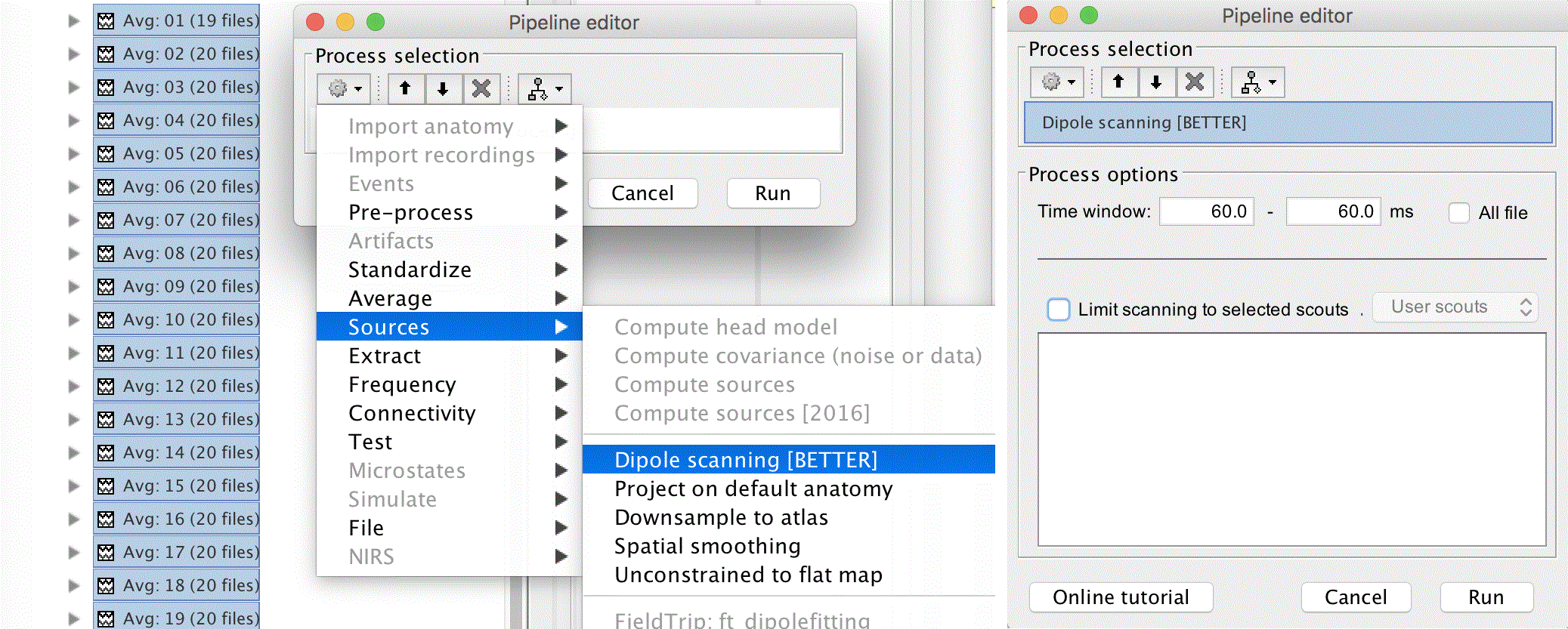
Dipole scanning
Select the set of 32 averages and drop them into the process box again. Switch to process sources and click the cog in the pipeline editor. Select Sources > Dipole Scanning [BETTER]. Set the process options to look at 60ms only by changing the Time window to 60ms - 60ms, and clicking Run.
Next we should average all the dipole fits together. To do this, we can expand the tree of average files and select Avg: 01 (19 files) | dipole scan through Avg: 32 (20 files) | dipole scan, then right click and select Merge dipoles. Alternatively, we can set Matlab to the study directory and use
FILES = dir('dipoles_fit*');
new = dipoles_merge({FILES.name});As an optional step, we can sign flip the dipoles in Matlab so that they are all pointing up for convenience. To do this, right click on Merge: 32 files and export to Matlab as the variable merge_dips. Then in Matlab use the following code:
for i = 1:32,
merge_dips.Dipole(i).Amplitude = merge_dips.Dipole(i).Amplitude*...
sign(merge_dips.Dipole(i).Amplitude(3));
end
merge_dips.Comment = [merge_dips.Comment ' | flipped'];
You can then right click on the channel file and click File > Import from Matlab, and bring merge_dips back into Brainstorm. With the comment added to the file, it should appear next to the previous merged dipoles, as Merge: 32 files | flipped.
Right click on the flippled dipole merge and click Diplay on MRI (3D), which brings up a figure showing the dipole locations. To compare these to the true locations in Matlab, you can run the following code:
[true_dipole_mat,true_loc,true_orient] = generate_phantom_neuromag_dips;
hold on
hq = quiver3(true_loc(1,:),true_loc(2,:),true_loc(3,:),...
true_orient(1,:),true_orient(2,:),true_orient(3,:));
hold off
set(hq,'color','r'),shgThe true dipole locations are now shown in red along with the estimated locations in blue: