|
Size: 7407
Comment:
|
← Revision 27 as of 2023-08-23 01:17:31 ⇥
Size: 6350
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| Half-day workshop, as part of the [[https://cuttingeeg2021.org/|CuttingEEG conference]]. This session will introduce new features for SEEG analysis: iEEG-BIDS specification, MRI segmentation with CAT12, MRI/CT coregistration and non-linear MNI normalization with SPM12, management of anatomical atlases, interactive marking of SEEG electrodes, automatic SEEG contact labelling based on volume and surface parcellations, epileptogenicity mapping, advanced FEM modeling including DTI-based constraints with DUNEuro. Participants will learn how to access these new tools from the Brainstorm environment on their personal laptops. |
Half-day workshop, as part of the [[https://cuttingeeg2021.org/|CuttingEEG conference]]. This session will introduce new features for SEEG analysis. Participants will learn how to access these new tools from the Brainstorm environment on their personal laptops. |
| Line 17: | Line 15: |
| <<HTML(<TR><TD>)>>'''Slides'''<<HTML(</TD><TD>)>>Introduction <<HTML(</TD></TR>)>> | <<HTML(<TR><TD>)>>'''Documents'''<<HTML(</TD><TD>)>>[[https://neuroimage.usc.edu/resources/cuttingeeg2021_brainstorm_slides.pdf|Introduction slides]] | [[https://neuroimage.usc.edu/resources/cuttingeeg2021_brainstorm_walkthrough.pdf|Tutorial walkthrough]] <<HTML(</TD></TR>)>> |
| Line 21: | Line 19: |
| == Program == | == Program (TBA) == |
| Line 23: | Line 21: |
Detailed program coming soon. <<HTML(<!-- )>> |
|
| Line 32: | Line 26: |
| <<HTML(<TR><TD>)>>9:15-9:45<<HTML(</TD><TD>)>>'''Lecture: Brainstorm overview<<BR>>''' | <<HTML(<TR><TD>)>>9:15-9:45<<HTML(</TD><TD>)>>'''Lecture: Brainstorm overview''' |
| Line 34: | Line 28: |
| Software structure, typical data workflow <<HTML(</TD></TR>)>> | Software structure, typical data workflow |
| Line 36: | Line 30: |
| <<HTML(<TR><TD>)>>9:45-10:30<<HTML(</TD><TD>)>>'''Anatomy''' | Presentation of the example dataset (BIDS-iEEG) <<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>9:45-10:45<<HTML(</TD><TD>)>>'''Anatomy''' |
| Line 42: | Line 40: |
| Anatomical parcellations |
|
| Line 44: | Line 44: |
| <<HTML(<TR><TD>)>>10:30-11:30<<HTML(</TD><TD>)>>'''SEEG recordings''' | <<HTML(<TR><TD>)>>10:45-11:45<<HTML(</TD><TD>)>>'''SEEG recordings''' |
| Line 52: | Line 52: |
| Projecting SEEG recordings on MRI and cortex surface | Anatomical labelling of SEEG contacts |
| Line 54: | Line 54: |
| <<HTML(<TR><TD>)>>11:30-12:15<<HTML(</TD><TD>)>>'''Epileptogenicity maps''' Importing epochs of interest in the database |
<<HTML(<TR><TD>)>>11:45-12:15<<HTML(</TD><TD>)>>'''Advanced topics - Based on participants' requests''' |
| Line 60: | Line 58: |
| Computation of epileptogenicity maps: Localization of the seizure onset zone | Epileptogenicity maps: Localization of the seizure onset zone and propagation pathways |
| Line 62: | Line 60: |
| Estimation of the propagation pathways | FEM demo: MRI segmentation, tissue anisotropy with DTI, FEM forward modeling Open discussion with Bainstorm users |
| Line 68: | Line 68: |
| <<BR>><<BR>> | <<BR>> |
| Line 72: | Line 72: |
| Setting up the software environment is an important part of the training and is a good preparation step to this workshop. Follow the instructions below if you are insterested in following the hands-on session on your own laptop. |
|
| Line 80: | Line 78: |
| Before coming to the workshop, you need to download the software and the tutorial dataset from the Brainstorm website (110Mb). Additionally, the epileptogenicity analysis is based on SPM12 (100Mb). To streamline troubleshooting during the session, please save all the downloaded files '''__on your Desktop__'''. | Before coming to the workshop, you need to download the software and the tutorial dataset from the Brainstorm website (200Mb). To streamline troubleshooting during the session, please save all the downloaded files '''__on your Desktop__'''. |
| Line 82: | Line 80: |
| 1. From the Brainstorm [[http://neuroimage.usc.edu/bst/download.php|Download]] page, log in or create a Brainstorm account | 1. Go to the [[http://neuroimage.usc.edu/bst/download.php|Download]] page, complete the quick and free registration process. 1. If you don't receive the confirmation email, please check your '''spam folder'''. |
| Line 84: | Line 84: |
| * '''Brainstorm software''': <<HTML(<FONT color=red>)>>brainstorm_YYMMDD.zip<<HTML(</FONT>)>> (70 Mb) * '''Tutorial dataset''': <<HTML(<FONT color=red>)>>workshop_cuttingeeg.zip<<HTML(</FONT>)>> (41 Mb) |
* '''Brainstorm software''': <<HTML(<FONT color=red>)>>brainstorm_YYMMDD.zip<<HTML(</FONT>)>> (source+binary, 90 Mb) * '''Tutorial dataset''': <<HTML(<FONT color=red>)>>workshop_cuttingeeg.zip<<HTML(</FONT>)>> (162 Mb) |
| Line 87: | Line 87: |
| 1. Download SPM12 from the UCL website, free registration required:<<BR>>http://www.fil.ion.ucl.ac.uk/spm/software/download/ 1. Unzip the three downloaded files on your desktop, then delete them |
1. Unzip the two downloaded files on your desktop, then delete them |
| Line 90: | Line 89: |
| 1. Final check: you should have now 4 folders on your desktop: | 1. Final check: you should have now 3 folders on your desktop: |
| Line 94: | Line 93: |
| * '''spm12''': SPM toolbox, required for the epileptogenicity analysis 1. Run Brainstorm, make sure it works correctly on your computer (read the next section) 1. Tell Brainstorm where you unzipped SPM12: Menu File > Edit preferences > SPM toolbox. |
1. Run Brainstorm, make sure it works correctly on your computer (see next sections) |
| Line 99: | Line 96: |
| ==== With Matlab ==== 1. Matlab versions >= 2014b are faster and produce nicer graphics: consider upgrading if possible |
If you don't have a Matlab license, you can get a temporary one for the duration of the conference: https://cuttingeeg2021.lpl-aix.fr/t/matlab-installation-licenses-available/151 ==== With Matlab (2014b minimum) ==== |
| Line 102: | Line 100: |
| Line 105: | Line 104: |
| 1. When asked for the Brainstorm database folder, pick the "brainstorm_db" you have just created 1. Tell Brainstorm where you unzipped SPM12: Menu File > Edit preferences > SPM toolbox. <<BR>>No need to add SPM to the Matlab path either, Brainstorm will do it when needed. |
1. When asked for the database folder, pick the "brainstorm_db" you have just created 1. '''Install SPM12''': Menu Plugins > spm12 > Install. |
| Line 109: | Line 108: |
| . The Brainstorm distribution includes a stand-alone version, created with the Matlab Compiler, which can be used without any Matlab license. However, compiled code cannot execute Matlab scripts that are not compiled, which prevents the use of external toolboxes (SPM, FieldTrip).<<BR>><<BR>>The last part of this hands-on session will rely on functions from the SPM toolbox, therefore if you use the compiled version, you wouldn't be able to follow the session until the end on your laptop. If you still want to run the compiled version of Brainstorm on your laptop, please refer to the main [[https://neuroimage.usc.edu/brainstorm/Installation|installation page]]. | 1. Install the Matlab Runtime '''R2020a (9.8)''': [[https://www.mathworks.com/products/compiler/matlab-runtime.html|Mathworks website]] 1. Run the program in brainstorm3/bin/R2020a/ * Windows: Double-click on ''brainstorm3.bat'' * MacOS: Double-click on ''brainstorm3.command ''and wait for instructions * Linux: From a terminal, run:<<BR>>''cd brainstorm3/bin/R2020a/<<BR>>'' ''./brainstorm3.command '' 1. For troubleshooting, see the [[https://neuroimage.usc.edu/brainstorm/Installation#Start_Brainstorm|Installation]] page. |
| Line 112: | Line 116: |
| In order to check if Brainstorm works properly or your computer, please follow these instructions: | To make sure Brainstorm works properly or your computer: |
| Line 114: | Line 118: |
| * Create a new protocol: Menu File > New protocol.<<BR>>Set the name to "Test", don't change the other default options. * In the folder "Default anatomy", double-click successively on the files: MRI, head, cortex_15002V * It should open the figures below, make sure they work correctly: * Click in the MRI Viewer to move the slices * Click+move your mouse in the 3D figure to rotate * Zoom in/out with the mouse wheel or the two-finger scroll on MacBook pads * The head surface should be transparent, with the brain visible through it {{attachment:test_bst.gif}} |
* Select the menu: '''Help > Workshop preparation'''. * Follow the instructions: when you see the 3D figure, make sure you see both a transparent head surface and a brain surface. Try to rotate the view with your mouse, and zoom in and out with the mouse wheel or two-finger pad scroll. <<BR>><<BR>> {{attachment:bst_test_2021.png}} |
| Line 125: | Line 123: |
<<HTML(--!>)>> |
Aix-en-Provence, France: October 4, 2021
Half-day workshop, as part of the CuttingEEG conference. This session will introduce new features for SEEG analysis. Participants will learn how to access these new tools from the Brainstorm environment on their personal laptops.
General information
| Where | Cube classrooms |
| When | Monday October 4th, 2021: 8:30-12:15 |
| Instructor | Francois Tadel |
| Audience | Users interested in analyzing sEEG/EEG/ECoG/MEG recordings using Brainstorm. Teaching in English. |
| Documents | Introduction slides | Tutorial walkthrough |
Program (TBA)
This session is based on a simplified version of the SEEG/Epileptogenicity tutorial.
| 8:30-9:15 | Onsite assistance in installing the material for the training session |
| 9:15-9:45 | Lecture: Brainstorm overview Software structure, typical data workflow Presentation of the example dataset (BIDS-iEEG) |
| 9:45-10:45 | Anatomy Database explorer MRI volumes, surfaces Anatomical parcellations Coregistration of pre- and post-implantation images |
| 10:45-11:45 | SEEG recordings Reviewing continuous SEEG recordings Montages and management of event markers Marking SEEG contacts on post-implantation image Anatomical labelling of SEEG contacts |
| 11:45-12:15 | Advanced topics - Based on participants' requests Time-frequency analysis: Identification of ictal HFO frequency bands Epileptogenicity maps: Localization of the seizure onset zone and propagation pathways FEM demo: MRI segmentation, tissue anisotropy with DTI, FEM forward modeling Open discussion with Bainstorm users |
Prepare your laptop for the workshop
Important notes
Working with MEG/EEG/SEEG recordings involves a lot of computational resources and large display windows. Therefore we recommend that you bring a laptop with a decent processing capacity, 4Gb RAM, a 64bit operating system, and a screen larger than 13".
You would add to your comfort by bringing an external mouse. Most of the manipulations are done with the mouse, and some involve an intensive use of the mouse wheel/scrolling.
Don't forget your power adapter!
Installation instructions
Before coming to the workshop, you need to download the software and the tutorial dataset from the Brainstorm website (200Mb). To streamline troubleshooting during the session, please save all the downloaded files on your Desktop.
Go to the Download page, complete the quick and free registration process.
If you don't receive the confirmation email, please check your spam folder.
Download the following files to your Desktop folder:
Brainstorm software: brainstorm_YYMMDD.zip (source+binary, 90 Mb)
Tutorial dataset: workshop_cuttingeeg.zip (162 Mb)
- If you already have Brainstorm on your laptop, make sure to update it before coming.
- Unzip the two downloaded files on your desktop, then delete them
Create a folder "brainstorm_db" on your Desktop
- Final check: you should have now 3 folders on your desktop:
brainstorm3: Program folder, with the source code and the compiled executable
brainstorm_db: Brainstorm database (empty)
workshop_cuttingeeg: Example dataset used during the training session
- Run Brainstorm, make sure it works correctly on your computer (see next sections)
Running Brainstorm for the first time
If you don't have a Matlab license, you can get a temporary one for the duration of the conference: https://cuttingeeg2021.lpl-aix.fr/t/matlab-installation-licenses-available/151
With Matlab (2014b minimum)
- Start Matlab
Do NOT add brainstorm3 folder to your Matlab path: this will be done automatically
- Go to the brainstorm3 folder
- Type "brainstorm" in the command window
- When asked for the database folder, pick the "brainstorm_db" you have just created
Install SPM12: Menu Plugins > spm12 > Install.
Without Matlab
Install the Matlab Runtime R2020a (9.8): Mathworks website
- Run the program in brainstorm3/bin/R2020a/
Windows: Double-click on brainstorm3.bat
MacOS: Double-click on brainstorm3.command and wait for instructions
Linux: From a terminal, run:
cd brainstorm3/bin/R2020a/
./brainstorm3.command
For troubleshooting, see the Installation page.
Make sure it works
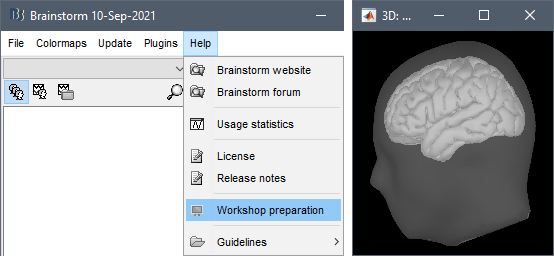
To make sure Brainstorm works properly or your computer:
Select the menu: Help > Workshop preparation.
Follow the instructions: when you see the 3D figure, make sure you see both a transparent head surface and a brain surface. Try to rotate the view with your mouse, and zoom in and out with the mouse wheel or two-finger pad scroll.
Troubleshooting
For any technical problem, please contact Francois Tadel ( francois.tadel@mcgill.ca )
