Live Brainstorm workshop and Q&A, Online: May 27, 2021
Two-hour online Brainstorm workshop and Q&A, live on Zoom.
General information
| Where | Zoom. |
| When | Thursday May 27, 2021: 10AM - 12PM EDT |
| Instructors | Sylvain Baillet (Montreal Neurological Institute / McGill, Canada) Raymundo Cassani (Montreal Neurological Institute / McGill, Canada) |
| Audience | Open to both beginners and advanced users. Bring your own questions. |
Program
This workshop is based on topics presented at detail in the get started tutorials.
Brainstorm overview
- Main user interface
- Software structure
- Typical data workflow
Anatomy data
- Database explorer
- MRI volumes and surfaces
- Default anatomy
Functional data
- Database explorer
- Reviewing recordings
- Visualization of recordings, montages and options
- Events and artifact removal with SSP
- Importing event-related data
- Forward model (head model)
- Inverse problem (source estimation) and normalization
- Visualization of sources, scouts and options
Process tab
- Process1 and Process2
- Overview of process categories
- Pipeline editor
- Process options and online help
- Pipeline as script
Q&A session
Prepare your laptop for the workshop (optional)
Due to time limitation this workshop is not hands-on, thus it is not required to install Brainstorm in your computer. However, if you decide to install Brainstorm and give try before the workshop, follow the instructions below.
Important notes
Working with MEG/EEG/SEEG recordings involves a lot of computational resources and large display windows. Therefore we recommend that you bring a laptop with a decent processing capacity, 4Gb RAM, a 64bit operating system, and a screen larger than 13".
You would add to your comfort by bringing an external mouse. Most of the manipulations are done with the mouse, and some involve an intensive use of the mouse wheel/scrolling.
Don't forget your power adapter!
Installation instructions
Before coming to the workshop, you need to download the software and the tutorial dataset from the Brainstorm website (330Mb). To streamline troubleshooting during the session, please save all the downloaded files on your Desktop.
From the Brainstorm Download page, log in or create a Brainstorm account
Download the following files to your Desktop folder:
Brainstorm software: brainstorm_YYMMDD.zip (60 Mb)
Tutorial dataset: sample_introduction.zip (330 Mb)
If you already have Brainstorm, make sure that the version was released after May 1st.
- Unzip the two downloaded files on your desktop
- Delete the two downloaded zip files
Create a folder "brainstorm_db" on your Desktop
- Final check: you should have now 3 folders on your desktop:
brainstorm3: Program folder, with the source code and the compiled executable
brainstorm_db: Brainstorm database (empty)
sample_introduction: Example dataset used during the training session
- Run Brainstorm, to make sure it works correctly on your computer (read the following section)
Running Brainstorm for the first time
With Matlab
Matlab versions >= 2014b are faster and produce nicer graphics: consider upgrading if possible
- Start Matlab
Do NOT add brainstorm3 folder to your Matlab path: this will be done automatically
- Go to the brainstorm3 folder
- Type "brainstorm" in the command window
- When asked for the Brainstorm database folder, pick the "brainstorm_db" you have just created
Without Matlab
- The instructions below are valid for the following operating systems:
Download the MCR R2015b (9.0) for your operating system: Mathworks website
- Install the MCR:
Windows: Double-click on the .exe and follow the instructions
MacOS: Click on the zip file to unzip it, then click on "InstallForMacOSX"
Linux: From a terminal, unzip .zip, then run ./install
- Run the program in brainstorm3/bin/R2015b/
Windows: Double-click on brainstorm3.bat
MacOS: Double-click on brainstorm3.command and wait for instructions
Linux: From a terminal, run:
cd brainstorm3/bin/R2015b/
./brainstorm3.command
- Troubleshooting for MacOS or Linux:
On recent versions of MacOS, you may get an error message "Application can't be opened because it is from an unidentified developer". This message would appear for all the programs that were not downloaded from the Apple app-store. To go around this verification: right-click on the application > Open, then click on the Open button.
From a terminal, make sure that the file "brainstorm3.command" is executable:
chmod a+x brainstorm3.commandIf nothing happens, run:
./brainstorm3.command MCR_DIR
MCR_DIR is the MCR 9.0 folder (ex: /Applications/MATLAB/MCR/v90)Try with another binary release: 2012b, 2013b, 2014b, 2016a, ...
- Troubleshooting for Windows:
Your current user may not have the necessary privileges. If you are an administrator for your computer, you can do the following: right-click on brainstorm3.bat > Run as administrator.
If you are not the administrator of your computer and Matlab or the MCR are not installed in the standard paths, Brainstorm may have trouble finding them. To specify manually the path of the MCR or Matlab folder, right-click on brainstorm3.bat > Edit. Fill the second line of the script (Example: @SET MATLABROOT="C:\Program Files\MATLAB\R2015b"), save the file, and try to execute it again.
- On Linux or MacOS, you may be asked to select the folder where you installed the MCR.
- When asked for the Brainstorm database folder, pick the "brainstorm_db" you have just created.
Make sure it works
In order to check if Brainstorm works properly or your computer, please follow these instructions:
Create a new protocol: Menu File > New protocol.
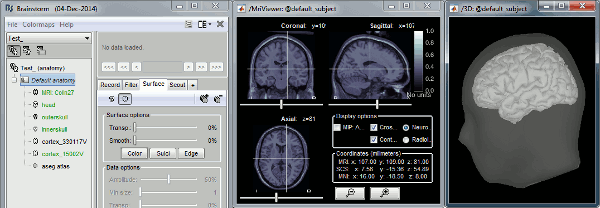
Set the name to "Test", don't change the other default options.- In the folder "Default anatomy", double-click successively on the files: MRI, head, cortex_15002V
- It should open the figures below, make sure they work correctly:
- Click in the MRI Viewer to move the slices
- Click+move your mouse in the 3D figure to rotate
Zoom in/out with the mouse wheel or the two-finger scroll on MacBook pads
- The head surface should be transparent, with the brain visible through it
Troubleshooting
For any technical problem, please contact Raymundo Cassani ( raymundo.cassani@mcgill.ca )
