|
Size: 3238
Comment:
|
Size: 9655
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Basic Electrophysiology in Brainstorm = ''Authors: Konstantinos Nasiotis'' |
= Brainstorm's Suite for Multi-unit Electrophysiology = ''Author: Konstantinos Nasiotis'' |
| Line 4: | Line 4: |
| <<TableOfContents(2,2)>> | {{attachment:intro.png||width="700"}} |
| Line 6: | Line 6: |
| {{attachment:intro.png}} | <<TableOfContents(3,2)>> |
| Line 9: | Line 9: |
| Invasive e-phys recordings aim in capturing the spiking activities of individual or small group of neurons. Since every spiking event is a very transient phenomenon (most spikes last around 1ms), the sampling rate of the recording system needs to be very high. Nowadays, acquisition systems can accommodate recordings from hundreds of channels from multiple multi-electrode arrays from different areas of the animal’s brain. A 10-minute recording that was used at the time of this tutorial from 224 channels (2 x 96 channel Utah arrays in PFC and TEO and one 32 channel depth electrode in the hippocampus) resulted to 10 min*60 sec*30,000 samples*224 channels* 2 bytes precision /1024 kb/1024 MB/1024 GB = ~7.5 GB. Since most recordings usually last much longer, there is a need for efficient data management and utilization of maximum processing power. | We feature a series of data analysis tools specifically dedicated to Invasive electrophysiology research (e-phys). We see this suite of expanded Brainstorm features as an opportunity for enabling a methodological continuum between (multi)cellular and systems level (with EEG and MEG) data analytics and facilitate the scientific dialogue and interactions between subdisciplines in the field. |
| Line 11: | Line 11: |
| Users that have Matlab’s parallel processing toolbox, will greatly benefit from the utilization of its properties. Most functions of the electrophysiology toolbox have a checkbox where users can enable parallelization of the processes. | Invasive electrophysiological recordings capture the spiking activity of individual or small groups of neurons. Since spiking events are transient phenomena (the typical duration of spikes is 1ms), the sampling rate of e-phys recording systems is typically much faster than for scalp EEG and MEG recordings. State-of-the-art acquisition systems can accommodate simultaneous recordings from hundreds of channels over several multi-electrode arrays positioned across the preparation, ultimately producing vast amounts of data. We foresee that Brainstorm will contribute to the efficient data management and processing when there is a need for efficient, open and reproducible data management and utilization of large quantities of data. Users that have Matlab’s parallel processing toolbox, will greatly benefit from the utilization of its properties. Most functions of the electrophysiology toolbox have a checkbox where users can enable parallelization of the processes. |
| Line 15: | Line 17: |
| All the function that will be described on this toolbox, are located in the __Electrophysiology__ processes tab. | All the function that will be described on this toolbox, are located in the ''Electrophysiology'' processes tab. |
| Line 19: | Line 21: |
| This tutorial uses a dataset collected from Dr. Christopher Pack’s lab (www.packlab.mcgill.ca), from a macaque monkey with a penetration chamber implanted in areas MT and MST. The recording probe consisted of 32 channels and 1 reference electrode. Users can download this tutorial data that will contain a converted Plexon acquisition system raw file (ytu288c-01_converted.bst), and an anatomy folder (Floyd MRI) that can all be imported to Brainstorm. The task for the animal was to maintain fixation on a target while a superflow stimulus was presented with different directions. The events from the stimulation system are saved along with pre-spike-sorted events. | Users that utilize this toolbox can cite: '''[[https://www.nature.com/articles/s41597-019-0242-z|Nasiotis, K., Cousineau, M., Tadel, F. et al. Integrated open-source software for multiscale electrophysiology. Sci Data 6, 231 (2019)]] ''''''''<<BR>>''''' '''''__[[https://nevronasio-my.sharepoint.com/:u:/g/personal/konstantinos_nevronas_io/ERm9f6sPf4tIrTZL9w2md5oB--lhKyGwkjBPJGpkHpoJCQ?e=JJYEiI|Tutorial Dataset]]__''''' This tutorial uses a dataset collected from Nardin Nakhla in Dr. Christopher Pack’s lab (www.packlab.mcgill.ca), from a macaque monkey with a penetration chamber implanted in areas MT and MST. The recording probe consisted of 32 channels and 1 reference electrode. Users can download this tutorial data that will contain a Plexon acquisition system raw file (ytu288c-01.plx), an events file (converted to Brainstorm format, that labels the experimental conditions appropriately), a channel positioning file and an anatomy folder (Floyd MRI) that can all be imported to Brainstorm. The task for the animal was to maintain fixation on a target while a superflow stimulus (dots that rotate, expand or translate in a partially coherent mode) was presented in one of 9 possible visual locations (as shown in the figure below). The events from the stimulation system are saved along with pre-spike-sorted events. {{attachment:stimulus.png||width="400"}} |
| Line 22: | Line 32: |
| The importation of raw e-phys data into Brainstorm follows the same procedure as it does for any other raw recording. Users firstly assign a name for their protocol/study, followed by the name of the subject/monkey, and sequentially can review their raw file. | {{attachment:importingRaw.png||width="700"}} The importing of raw e-phys data into Brainstorm follows the same procedure as it does for any other raw recording. Users firstly assign a name for their protocol/study, followed by the name of the subject/monkey, and sequentially can review their raw file. |
| Line 26: | Line 38: |
| '''''__Create Protocol__''''' | === Create protocol === * Create a new protocol: File > New protocol. * Give the Protocol name: ''Tutorial_e-Phys. '' * Default anatomy: ''No, use individual anatomy. '' * Default channel File: ''No, use one channel file per acquisition run.''<<BR>><<BR>> {{attachment:newProtocol.png}} * Press create, and this will create the new protocol. |
| Line 28: | Line 45: |
| * When a new study needs to be analyzed, users have to create a new protocol: File -> New Protocol. * Give the Protocol name: Tutorial_e-Phys. * For the anatomy set it to: No, use individual anatomy. |
=== Add subject === The next step will be to set the subject/monkey: * Right-click on the protocol created: Tutorial_e-Phys (subjects) * Select ''New subject'' * Make sure the tab ''Functional Data (sorted by subjects)'' is selected – select it under the Tutorial_e-Phys drop-down.<<BR>><<BR>> {{attachment:newSubject_new.png}} * A new window will appear that allows users to set the Subject name and to choose the individual anatomy and default channel file. Choose no for both default anatomy and default channel file. * Use subject Name: ''Floyd'' <<BR>><<BR>> {{attachment:FloydSubject.png}} * Once Save is selected, a new subject (Floyd) will be created in the database. === Import anatomy (Optional) === Users can now import the MRI if needed. Instructions on how to import the MRI can be found [[https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy|here]] and just use the Floyd MRI folder as the Freesurfer/anatomy folder. However, the analysis described on later steps (besides a few visualization functions) does not require an anatomical MRI. === Import recordings === Raw files can be imported by selecting the acquisition system used, and sequentially selecting the raw file. Just right-click on the Subject (Floyd) and then click on ''Review raw file.'' <<BR>> {{attachment:reviewRaw_main_window.png}} <<BR>> <<BR>><<BR>> A new window will pop-up that allows users to select the acquisition system and the raw file. Select EEG:Plexon (*.plx;.pl2) as the acquisition system and navigate to the folder the ytu288c-01.plx raw file is saved. <<BR>> {{attachment:selectRaw_popup.png||width="600"}} <<BR>> <<BR>> Once the procedure is complete, this will create a new folder ''(ytu288c-01)'' and two new files in the database: a channel file (''Plexon channels (33)'') and the ''Link to raw file''. These two files contain all the information from the header of the acquisition system. {{{#!wiki note Importing of this file might last a few minutes. This is not typically the case for files from other acquisition systems. .plx files suffer from inefficient data loading and can cause severe waiting times, especially when users are scrolling through the end of the recording. In order to avoid this, the .plx files are converted into .bst files (Brainstorm binary format) for an improved user experience. }}} {{attachment:importedRaw_main_window.png}} <<BR>> <<BR>> By double clicking on the Link to raw file, a viewer will open that allows the user to navigate through the recordings. This is a very easy way to inspect the recordings for artifacts or stimulation effects since only the selected part is loaded in memory on-the-fly, without importing anything in the database. Any events captured from the acquisition system are automatically loaded from the importer. <<BR>><<BR>> {{attachment:displayRaw_new.png||width="700"}} <<BR>><<BR>> It is important to note that some acquisition systems might have enabled their own automatic spike sorter. These events are automatically labeled with the convention that is used throughout the e-phys toolbox: “Spikes Channel ''ChannelLabel''”, where ChannelLabel is the Label of the electrode assigned by the acquisition system. If the acquisition system assigned the spikes’ waveforms into more than one cluster/neuron during this automatic clustering, the neuronal events would be (e.g. for an electrode that picked up spikes from 3 neurons): “Spikes Channel ChannelLabel |1|”, “Spikes Channel ChannelLabel |2|”, “Spikes Channel ChannelLabel |3|”. For more information, users can visit [[https://neuroimage.usc.edu/brainstorm/e-phys/ConvertToBrainstormEvents|this]] page. The events are comprised of the signals that the stimulation system has sent (Stim On, Start, Stop etc.) and the pre-spike-sorted events that the Plexon acquisition system can perform (Spikes Channel AD01, Spike Channel AD02 etc.). === Add additional events === In case users need to input events from a separate file, this can be easily done by clicking on the events panel: File > Add events from file. Several event filetypes are supported. Specifically for the tutorial dataset, the stimulus presentation events were coded from a software that was not triggering events on the acquisition system, so they need to be manually imported. Users should first double click on the Link to raw file (this activates the Record tab) and then the events can be added by selecting the file: ytu288c-01_converted_events.csv <<BR>> {{attachment:import_events_new.png||width="300"}} <<BR>> <<BR>> Select files of type: ''CSV test file'' and navigate to the folder that contains the tutorial files. <<BR>> <<BR>> {{attachment:select_events_new.png||width="600"}} <<BR>> <<BR>> This procedure will add 9 new types of events at the end of the events list: <<BR>> <<BR>> {{attachment:imported_events_new.png||width="400"}} ---------- At this stage, users can start performing their analysis based on the experimental events. <<EmbedContent("http://neuroimage.usc.edu/bst/get_prevnext.php?next=/e-phys/SpikeSorting")>> <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?e-phys/Introduction)>> |
Brainstorm's Suite for Multi-unit Electrophysiology
Author: Konstantinos Nasiotis
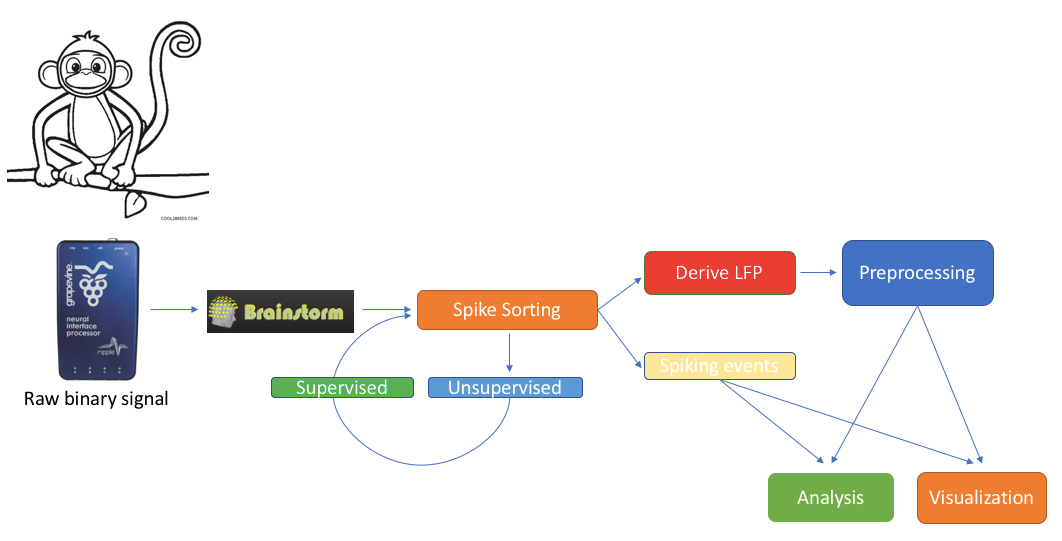
Contents
Introduction
We feature a series of data analysis tools specifically dedicated to Invasive electrophysiology research (e-phys). We see this suite of expanded Brainstorm features as an opportunity for enabling a methodological continuum between (multi)cellular and systems level (with EEG and MEG) data analytics and facilitate the scientific dialogue and interactions between subdisciplines in the field.
Invasive electrophysiological recordings capture the spiking activity of individual or small groups of neurons. Since spiking events are transient phenomena (the typical duration of spikes is 1ms), the sampling rate of e-phys recording systems is typically much faster than for scalp EEG and MEG recordings. State-of-the-art acquisition systems can accommodate simultaneous recordings from hundreds of channels over several multi-electrode arrays positioned across the preparation, ultimately producing vast amounts of data.
We foresee that Brainstorm will contribute to the efficient data management and processing when there is a need for efficient, open and reproducible data management and utilization of large quantities of data. Users that have Matlab’s parallel processing toolbox, will greatly benefit from the utilization of its properties. Most functions of the electrophysiology toolbox have a checkbox where users can enable parallelization of the processes.
Additionally, it is highly recommended for users that need to analyze large files, to select a large number for memory utilization on any processes that allow users to adjust the memory used, since the speed of the processes (especially the spike sorting procedures that use demultiplexing of the raw signal) can be greatly improved by the extra memory used.
All the function that will be described on this toolbox, are located in the Electrophysiology processes tab.
Any toolboxes embedded represent the work of their respective authors and need to be appropriately cited. Links to the papers that the toolboxes correspond to are located on each section.
Users that utilize this toolbox can cite:
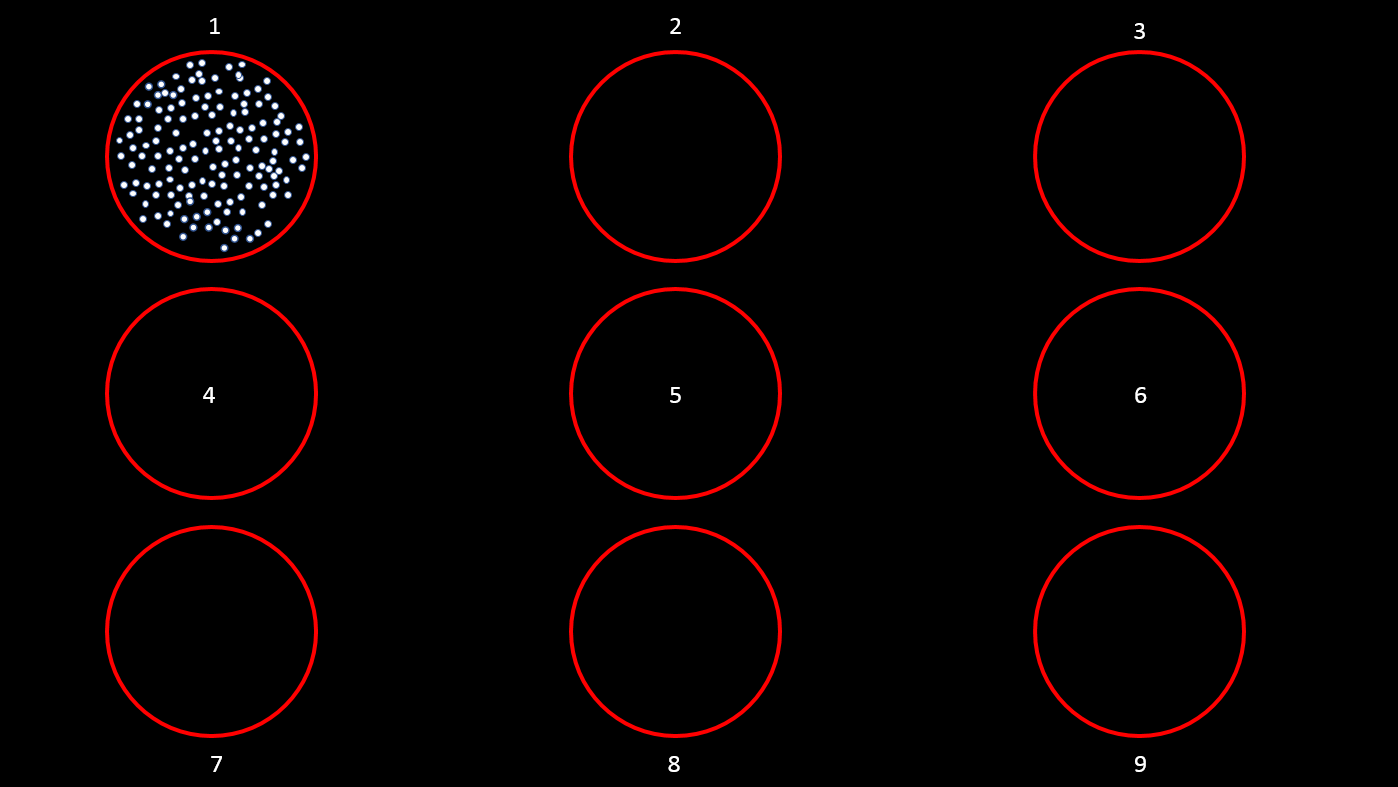
This tutorial uses a dataset collected from Nardin Nakhla in Dr. Christopher Pack’s lab (www.packlab.mcgill.ca), from a macaque monkey with a penetration chamber implanted in areas MT and MST. The recording probe consisted of 32 channels and 1 reference electrode. Users can download this tutorial data that will contain a Plexon acquisition system raw file (ytu288c-01.plx), an events file (converted to Brainstorm format, that labels the experimental conditions appropriately), a channel positioning file and an anatomy folder (Floyd MRI) that can all be imported to Brainstorm. The task for the animal was to maintain fixation on a target while a superflow stimulus (dots that rotate, expand or translate in a partially coherent mode) was presented in one of 9 possible visual locations (as shown in the figure below). The events from the stimulation system are saved along with pre-spike-sorted events.
Importing raw e-phys data
The importing of raw e-phys data into Brainstorm follows the same procedure as it does for any other raw recording. Users firstly assign a name for their protocol/study, followed by the name of the subject/monkey, and sequentially can review their raw file.
All these steps are briefly repeated here for the example e-Phys file for completeness.
Create protocol
Create a new protocol: File > New protocol.
Give the Protocol name: Tutorial_e-Phys.
Default anatomy: No, use individual anatomy.
Default channel File: No, use one channel file per acquisition run.
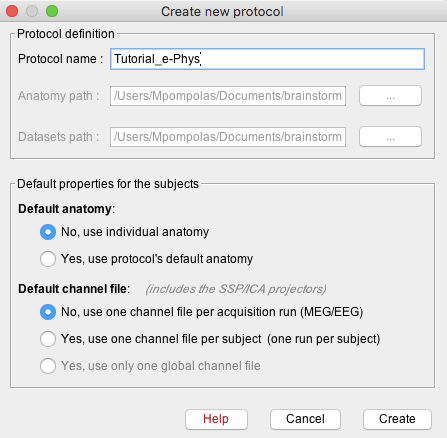
- Press create, and this will create the new protocol.
Add subject
The next step will be to set the subject/monkey:
- Right-click on the protocol created: Tutorial_e-Phys (subjects)
Select New subject
Make sure the tab Functional Data (sorted by subjects) is selected – select it under the Tutorial_e-Phys drop-down.
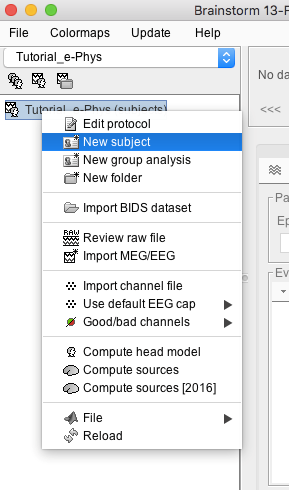
- A new window will appear that allows users to set the Subject name and to choose the individual anatomy and default channel file. Choose no for both default anatomy and default channel file.
Use subject Name: Floyd
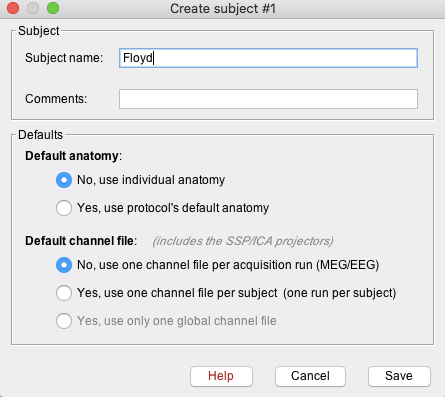
- Once Save is selected, a new subject (Floyd) will be created in the database.
Import anatomy (Optional)
Users can now import the MRI if needed. Instructions on how to import the MRI can be found here and just use the Floyd MRI folder as the Freesurfer/anatomy folder. However, the analysis described on later steps (besides a few visualization functions) does not require an anatomical MRI.
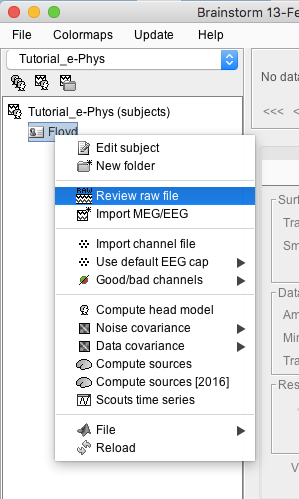
Import recordings
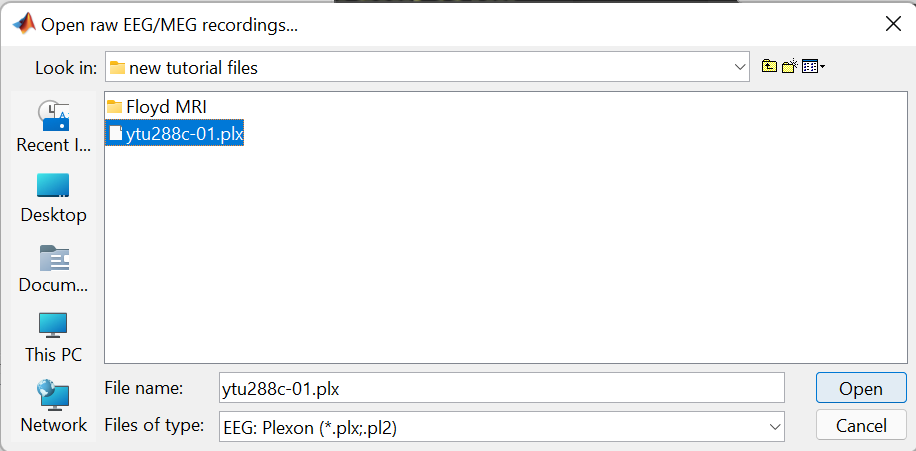
Raw files can be imported by selecting the acquisition system used, and sequentially selecting the raw file. Just right-click on the Subject (Floyd) and then click on Review raw file.
A new window will pop-up that allows users to select the acquisition system and the raw file. Select EEG:Plexon (*.plx;.pl2) as the acquisition system and navigate to the folder the ytu288c-01.plx raw file is saved.
Once the procedure is complete, this will create a new folder (ytu288c-01) and two new files in the database: a channel file (Plexon channels (33)) and the Link to raw file. These two files contain all the information from the header of the acquisition system.
Importing of this file might last a few minutes. This is not typically the case for files from other acquisition systems. .plx files suffer from inefficient data loading and can cause severe waiting times, especially when users are scrolling through the end of the recording. In order to avoid this, the .plx files are converted into .bst files (Brainstorm binary format) for an improved user experience.
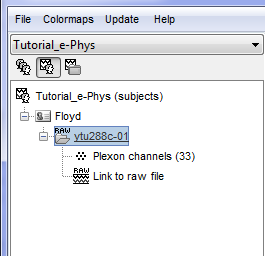
By double clicking on the Link to raw file, a viewer will open that allows the user to navigate through the recordings. This is a very easy way to inspect the recordings for artifacts or stimulation effects since only the selected part is loaded in memory on-the-fly, without importing anything in the database. Any events captured from the acquisition system are automatically loaded from the importer.
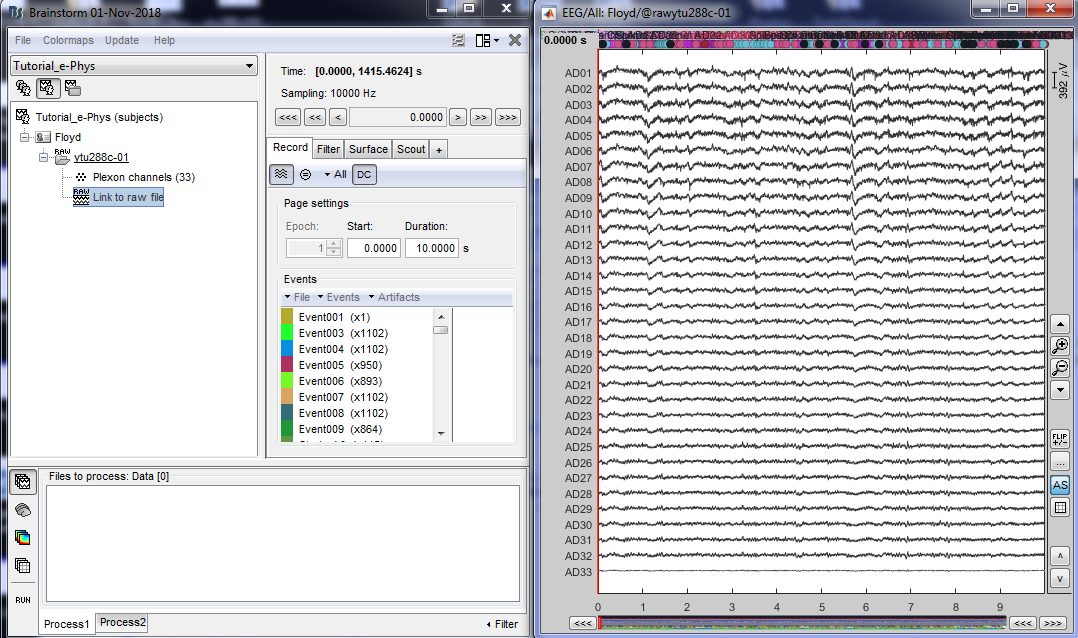
It is important to note that some acquisition systems might have enabled their own automatic spike sorter. These events are automatically labeled with the convention that is used throughout the e-phys toolbox:
“Spikes Channel ChannelLabel”, where ChannelLabel is the Label of the electrode assigned by the acquisition system. If the acquisition system assigned the spikes’ waveforms into more than one cluster/neuron during this automatic clustering, the neuronal events would be (e.g. for an electrode that picked up spikes from 3 neurons): “Spikes Channel ChannelLabel |1|”, “Spikes Channel ChannelLabel |2|”, “Spikes Channel ChannelLabel |3|”.
For more information, users can visit this page.
The events are comprised of the signals that the stimulation system has sent (Stim On, Start, Stop etc.) and the pre-spike-sorted events that the Plexon acquisition system can perform (Spikes Channel AD01, Spike Channel AD02 etc.).
Add additional events
In case users need to input events from a separate file, this can be easily done by clicking on the events panel: File > Add events from file. Several event filetypes are supported.
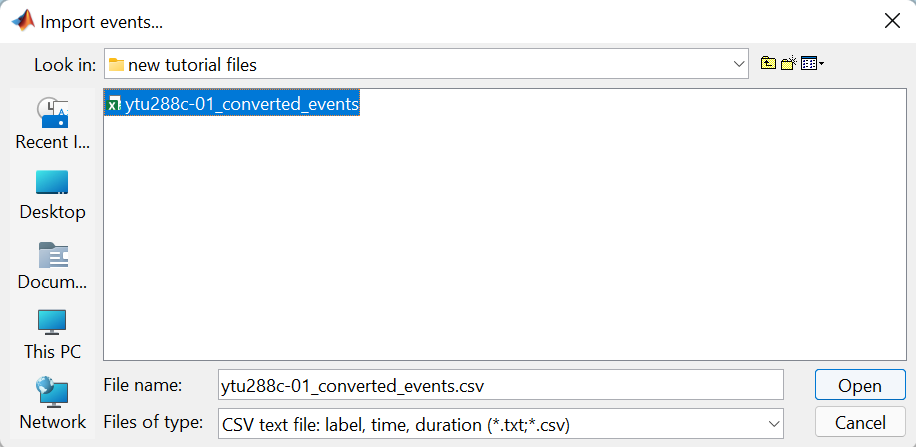
Specifically for the tutorial dataset, the stimulus presentation events were coded from a software that was not triggering events on the acquisition system, so they need to be manually imported. Users should first double click on the Link to raw file (this activates the Record tab) and then the events can be added by selecting the file: ytu288c-01_converted_events.csv
Select files of type: CSV test file and navigate to the folder that contains the tutorial files.
This procedure will add 9 new types of events at the end of the events list:
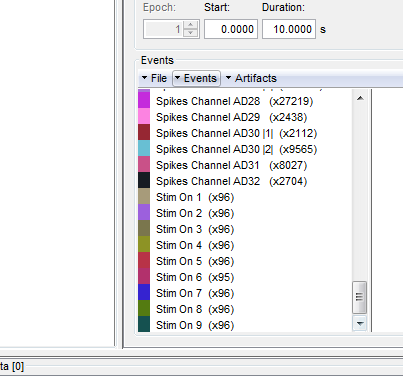
At this stage, users can start performing their analysis based on the experimental events.
