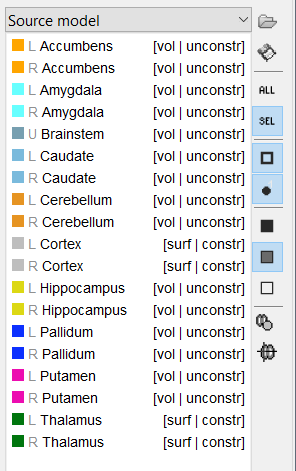
Hi Martin/Francois,
I am working with mixed models for the first time in a while and have noticed a few curiosities. (Brainstorm is updated to 13 April 2020; Matlab 2019a)
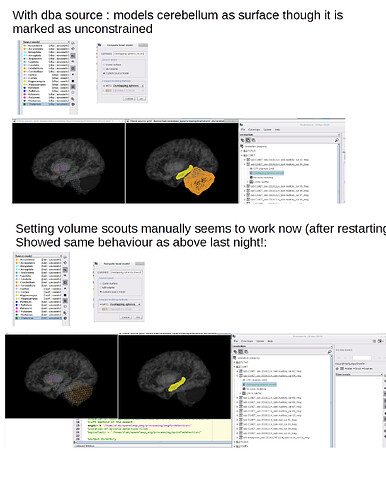
1. Forward mixed model assigns cerebellum to surface rather than volume, and not cortex.
Steps:
- under Anatomy tab, I create a source model option atlas will all structures (which include cortex and subcortical regions) set to deep brain (dba). The structures correctly default to cortex as surface (constr), subcortical structures as volume (unconstrained).
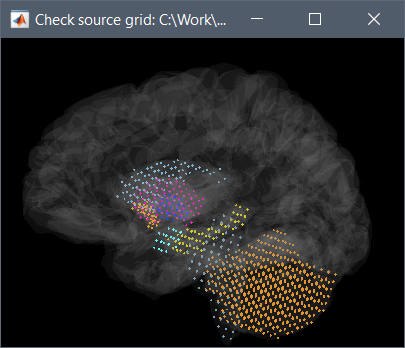
- under the Functional tab, I create a mixed overlapping spheres head model. Looking at the grid (Check source grid (volume) ), the cerebellum is not included in the volume portion, though the other non-cortical structures are. Looking at the surfaces, only hippocampus and cerebellum are represented (not pallidum and not cortex)
- Troubleshooting attempt: if I go back to the anatomy tab Source Model atlas and set everything to Volume, except for cortex and thalamus to surface, and repeat the overlapping spheres head model, it does not correct the problem - cerebellum still appears to be a surface and cortex doesn't exist at all, from which I conclude it is not the dba defaults but rather something more general.
2. Forward mixed model calculation allows you to set different grid sizes, but ignores the argument.
Steps:
- for mixed models, the option to change grid size can be accessed if you run the forward model via the Process 1 tab. (Compute head model > Custom source model, MRI volume grid: Edit: Regular grid (isotropic), grid resolution ....)
- however, changing the value doesn't change the outcome, which seems to default to a 5mm iso grid regardless of the numbers you put in. (This grid size is inadequate for some of the smaller structures)
- Troubleshooting attempt: I redid the overlapping spheres head model with the Generate from Cortical Surface option and visualized it, and it still looks like a 5mm iso grid only on the brainstem and subcortical regions.
3. Importing volume masks on mixed models (I don't know if this is a bug or just not possible yet)
- I have a subject-space subcortical volume-based mask that works nicely on full-volume models. If I read it into a mixed model, it doesn't give an error and adds an atlas with scouts, but I don't see anything appear on the brain visualization.
- Troubleshooting attempt: I tried to read it in on a full-volume model, export the scouts to matlab, and read them in on the mixed model that had the same iso grid. They appear, but not in the right place. Is there any way to read in subcortical masks on mixed models? (That would be very useful!)
Best regards,
Emily