In the “sources” memu, there is a submenu “downsampling to atlas”.
It is a wonderful function I have to say.
However, I have a question.
When I write papers, how to explain such operations, its implication and significiance?
Would you like to give me some literatures to support such function?
Best wishes,
Junpeng
Hi Junpeng,
This approach “downsampling to atlas” was implemented to simplify the source models before a full brain connectivity analysis. Computing all the pairs of connectivity measures (15000 x 15000) was not realistic on most computers, so we explored the options available to subdivide the source maps in regions of interest (between 50 and 300) to produce more accessible NxN connectivity matrices.
But the problem of how to perform the parcellation of the cortex before running this “downsampling to atlas” is a very complicated one: all the regions should have similar sizes (using the anatomical atlases from FreeSurfer introduce important biases in the signals because some regions are large and other very small), and they should follow the functional maps (but then, how to get the same parcellation across subjects?). We tried a few things but there were too many unknowns and we didn’t have enough resources to explore further this approach.
We don’t use much this menu “downsampling to atlas” ourselves: either we work with full brains (15000 sources) or a limited number of regions of interests based on the hypotheses of the experiment (manually defined or from a FreeSurfer anatomical atlas), depending on the questions. I don’t have any good reference to cite to justify this method.
Robert Oostenveld and the FieldTrip team explored further this approach when designing the analysis pipeline for the MEG part of the Human Connectome Project (http://www.humanconnectome.org/documentation/HCP-pipelines/meg-pipeline.html). For more information and references to cite, you could maybe post your question on the FieldTrip mailing list (http://www.fieldtriptoolbox.org/discussion_list).
Cheers,
Francois
1 Like
Hello,I am using Human connectome project database for MEG signals which has 248 channel. So, I am going to use some predefined atlas of brainstorm in order to have maximally 20 ROI, I would like to know what are my steps to doing so?
Thanks
Hello,
What are the steps you don’t know how to perform?
Please refer to the introduction tutorials (follow them if you haven’t yet) to indicate which section of the pipeline you need help with.
Please note that we gave up trying to use the HCP-MEG data because the authors didn’t release the digitized head points, which are indispensable to compute an accurate registration between the MRI and the MEG.
Cheers,
Francois
Thanks Francois, It was really helpful, I am going to use the HCP MEG data in inverse problem. I recently read the article by G.L.Colclough about “How reliable are MEG resting-state connectivity metrics?” and they used HCP dataset and then registered into the standard co-ordinate space of the Montreal neuroimging , I am wondering if there is way to do what they did in brainstorm?
Thanks
Hello,
Yes, it is possible to project on an MNI template the sources computed for the individual subject (http://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects) and them to compute connectivity metrics (no tutorial yet, but coming soon).
However it is difficult to use the HCP data because the head shape points are not provided in this database, therefore it is difficult to register accurately the MRI with MEG with our standard processing pipeline. The transformation they used in the FieldTrip scripts are distributed with the data but we haven’t written yet the routines to import them automatically.
Francois
Hello,
is there any way to import the computed eloreta result by fieldtrip based on human connectome project into BRAINSTORM for parcellation?
Thanks
Hello
Right now it is not possible to import these files, but it might be added at some point.
I’m currently working on adding the support of the FieldTrip functions for computing forward and inverse solutions, so that we can use them in Brainstorm. But I’m not sure how feasible it will be to import the FieldTrip source maps in Brainstorm.
Check regularly the list of software updates: http://neuroimage.usc.edu/brainstorm/Updates
It is basically a problem of conversion of Matlab structures. If you are very comfortable with this, you can try to do it yourself. You can compute some volume source models in Brainstorm, then try to replace the fields in the Brainstorm files with the data you want to use from the FieldTrip structures.
Documentation of the source files: http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#On_the_hard_drive
Volume head models: http://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource
Import/export structures: http://neuroimage.usc.edu/brainstorm/Tutorials/Scripting#Custom_processing
Francois
Dear Brainstorm team,
We have averaged (across each condition) event-related data for which we computed 4 unconstrained sLoretta kernels (1 for each of the three conditions and 1 for the average across all conditions).
We would now like to calculate/return to a time series. We are trying to
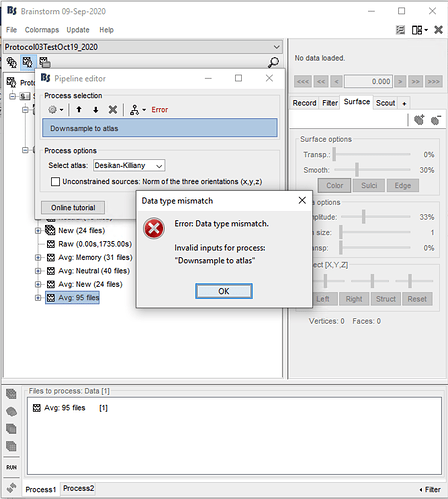
- Downsample to atlas (Desikan-Killiany)
- Extract > Scout timeseries
However, we get an error in the first step when downsampling to atlas (screenshots attached): "Error: Data type mismatch. Invalid inputs for process: 'Downsample to atlas' "
Please let us know if there is an error in our steps or if there is a different way to get the timeseries from the sLoretta kernel.
Thank you very much,
Manda