Thanks a lot. At first, it made a lot of sense to me.
But there is one thing i don't get. If i run mri_info (function from freesurfer), then i see the vox2ras transformation just fine:
(base) edelaire@perf-imglab07:/NAS/home/edelaire/Documents/data/NeuroDot/2014_Nph_Data_Share/template$ mri_info mni152nl_T1_on_333_nifti.nii
Volume information for mni152nl_T1_on_333_nifti.nii
Primary Slice Direction: axial
voxel to ras transform:
3.0000 0.0000 0.0000 -70.5000
0.0000 3.0000 0.0000 -105.0000
0.0000 0.0000 3.0000 -60.0000
0.0000 0.0000 0.0000 1.0000
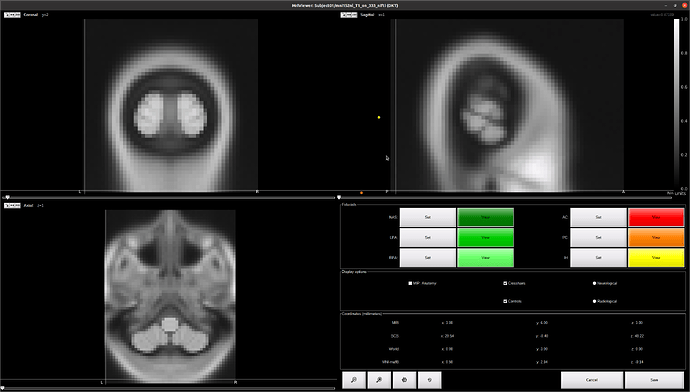
Can it be an issue when loading MRI in brainstorm ?
I tried also on the 1x1x1 and it seems present too:
(base) edelaire@perf-imglab07:/NAS/home/edelaire/Documents/data/NeuroDot/2014_Nph_Data_Share/template$ mri_info mni152nl_T1_on_111_nifti.nii
Volume information for mni152nl_T1_on_111_nifti.nii
type: nii
dimensions: 176 x 208 x 176
voxel sizes: 1.000000, 1.000000, 1.000000
type: FLOAT (3)
fov: 176.000
dof: 1
xstart: -88.0, xend: 88.0
ystart: -104.0, yend: 104.0
zstart: -88.0, zend: 88.0
TR: 0.00 msec, TE: 0.00 msec, TI: 0.00 msec, flip angle: 0.00 degrees
nframes: 1
PhEncDir: UNKNOWN
FieldStrength: 0.000000
ras xform present
xform info: x_r = 1.0000, y_r = 0.0000, z_r = 0.0000, c_r = 0.0000
: x_a = 0.0000, y_a = 1.0000, z_a = 0.0000, c_a = -19.0000
: x_s = 0.0000, y_s = 0.0000, z_s = 1.0000, c_s = 13.0000
Orientation : RAS
Primary Slice Direction: axial
voxel to ras transform:
1.0000 0.0000 0.0000 -88.0000
0.0000 1.0000 0.0000 -123.0000
0.0000 0.0000 1.0000 -75.0000
0.0000 0.0000 0.0000 1.0000
voxel-to-ras determinant 1
ras to voxel transform:
1.0000 0.0000 0.0000 88.0000
0.0000 1.0000 0.0000 123.0000
0.0000 0.0000 1.0000 75.0000
0.0000 0.0000 0.0000 1.0000
So i looked a bit, at the import function and it seems that both sForm and qForm are defined and that qForm (wrong) is loaded when the correct one seems to be in sForm
K>> nifti.sform
ans =
3.0000 0 0 -70.5000
0 3.0000 0 -105.0000
0 0 3.0000 -60.0000
0 0 0 1.0000
K>> nifti.qform
ans =
61.6615 0 0 0
0 61.7209 0 0
0 0 61.5116 0
0 0 0 1.0000
If i force brainstorm to use the sform, then it's working, without calling SPM for realignment:
Is that an issue with the MRI file; or with Brainstorm reading function ?