Hello,
This post is the continuation of Aligned MRI outside brainstorm are no longuer aligned after importation
Additionally to the fMRI data, nicely packed in nifty. The group we are working with sent us some additional data as .mat file; that I need to import and visualize on top of the MRI imported in the other topic.
Data:
The data can be downloaded here : https://drive.google.com/file/d/1cIL3gAukUo41JKUTtn_7inw_Pprq3Y6O/view?usp=sharing
sMriRef is from brainstorm and corresponds to the T1 imported after FreeSurfer and correspond to my target MRI.
sMriSrc is the data that I received and that I tried to format as much as possible as Brainstorm.
Here is what I know:
sMriSrc is aligned to the subject MRI ( I don't have access to it) but they computed the transformation to align the subject MRI to the MNI template (sMriRef).
This transformation is stored under: sMriSrc.info.tissue.affine
What I tried:
I tried to copy from mri_resclice code , using
% Transformation subject => MNI
TransfSrc = sMriSrc.info.tissue.affine;
and
TransfRef = [sMriRef.SCS.R, sMriRef.SCS.T; 0 0 0 1];
with the transformation of the grid being:
allGrid = inv(TransfSrc) * TransfRef * allGrid;
function sMriReg = reslice_mri(sMriSrc, sMriRef)
% Reslice the MRI to target MRI. Based on mri_reslice
sMriSrc.Voxsize = [sMriSrc.info.tissue.dim.mmx, sMriSrc.info.tissue.dim.mmy, sMriSrc.info.tissue.dim.mmz];
% Transformation subject => MNI
TransfSrc = sMriSrc.info.tissue.affine;
TransfRef = [sMriRef.SCS.R, sMriRef.SCS.T; 0 0 0 1];
% ===== INTERPOLATE MRI VOLUME =====
% Original position vectors (WATCH OUT FOR THE X/Y PERMUTATION OF MESHGRID!)
X1 = (0:size(sMriSrc.Cube,1)-1) + 0.5;
Y1 = (0:size(sMriSrc.Cube,2)-1) + 0.5;
Z1 = (0:size(sMriSrc.Cube,3)-1) + 0.5;
% Reference position vectors
X2 = (0:size(sMriRef.Cube,1)-1) + 0.5;
Y2 = (0:size(sMriRef.Cube,2)-1) + 0.5;
Z2 = (0:size(sMriRef.Cube,3)-1) + 0.5;
% Mesh grids
[Xgrid2, Ygrid2, Zgrid2] = meshgrid(Y2, X2, Z2);
% Apply final transformation: reference MRI => common space => original MRI
allGrid = [Ygrid2(:)' .* sMriRef.Voxsize(1); ...
Xgrid2(:)' .* sMriRef.Voxsize(2); ...
Zgrid2(:)' .* sMriRef.Voxsize(3); ...
ones(size(Xgrid2(:)))'];
allGrid = inv(TransfSrc) * TransfRef * allGrid;
Xgrid2 = reshape(allGrid(2,:), size(Xgrid2));
Ygrid2 = reshape(allGrid(1,:), size(Ygrid2));
Zgrid2 = reshape(allGrid(3,:), size(Zgrid2));
% OPTION #2: Cubic interp, very similar results, much faster
n4 = size(sMriSrc.Cube,4);
newCube = cell(1,n4);
for i4 = 1:n4
newCube{i4} = single(interp3(...
Y1 .* sMriSrc.Voxsize(2), ...
X1 .* sMriSrc.Voxsize(1), ...
Z1 .* sMriSrc.Voxsize(3), ...
double(sMriSrc.Cube(:,:,:,i4)), Xgrid2, Ygrid2, Zgrid2, 'cubic', 0));
end
newCube = cat(4, newCube{:});
% Save output
sMriReg = sMriRef;
sMriReg.Cube = newCube;
sMriReg.Comment = 'Imported volume';
end
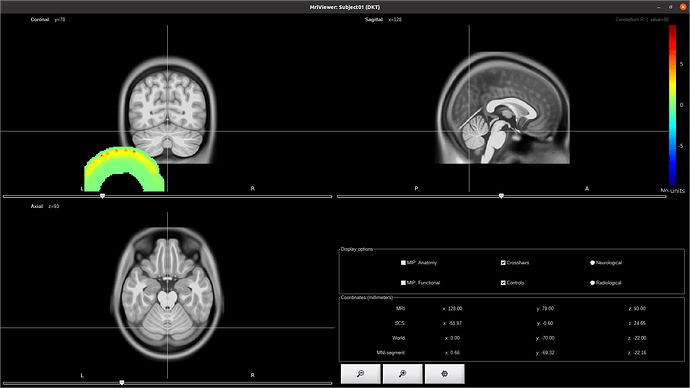
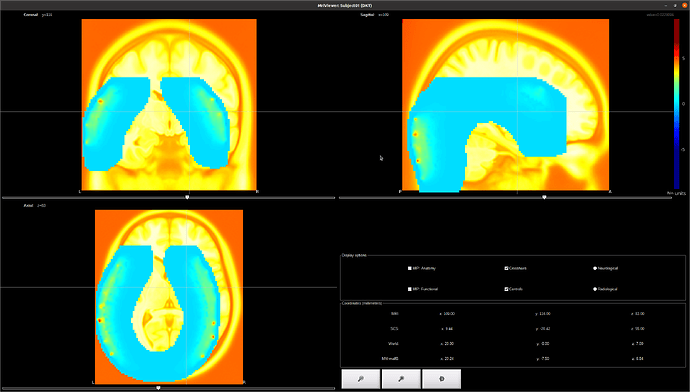
The issue is that it is not aligned :
Would you be able to help me on applying the transformation ? The map is supposed to cover the occipital / temporal lobe of the MRI.
I am wondering if the volume is actually saved as x/y/z or if there is some inversion... but I am not sure on how to check that
Edouard