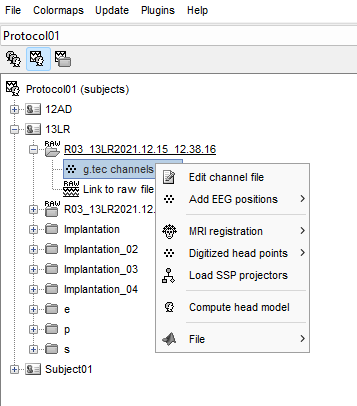
I changed channel group as SEEG for all electrodes and I renamed all the electrode groups by letters ascending alphabetical order, but when I go to edit electrodes there is only 1 electrode letter E.
Your screen capture shows that you didn't edit the channel names. With these channel names, indeed, it defines 1 SEEG shaft with 205 contacts on it.
Don't you have access to the implantation scheme of the neurosurgeon?
I guess they don't use these labels E001..E205 for the clinical evaluation.
Maybe something like:
E001 => A1 or A12
E002 => A2 or A11
E012 => A12 or A1
E013 => B1 or B16
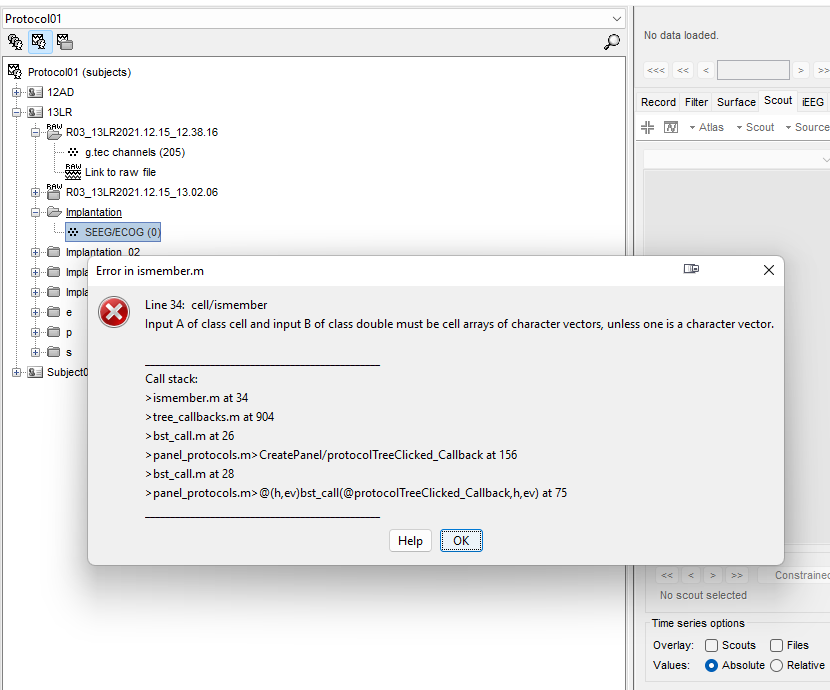
When you open for display the channel file for the first time, Brainstorm parses the channel names and creates groups and intracranial electrodes automatically (IntraElectrodes in the channel file: https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#On_the_hard_drive).
If you later edit the channel names/types/groups, the IntraElectrodes doesn't get updated automatically. To force updating it: in the iEEG tab, delete ALL the electrodes in the list, close everything (big cross in the toolbar), save the changes, and open the channel file again. It would re-create the IntraElectrodes with the new information from the channel file.
If you are not comfortable with the manipulation of the channel files, I recommend you start by following step by step the three EEG/SEEG/ECOG tutorials using the example dataset before processing your own data:
I think I'm missing the point of the channel group
One channel group = one IntraElectrode created at the first time the channel file is loaded for display (or when there is no IntraElectrode defined, i.e. when it all the Electrodes has been deleted)
I'm using postop CT SPM reslice for MRI registration to set the electrode tip and entry points. Is it better to use MNI normalized SPM of postop CT?
Never use volumes normalized to MNI space when working with SEEG. Localize the electrodes only on volumes in the original patient space.
Is Cat12 or Brainsuite preferred for surface generation?
No formal comparison of the surfaces generated with the two programs has been published to my knowledge, therefore we can't give you any recommendation.
CAT12 is much faster, indeed.
Is iEEG atlas labeling of contacts meant to "find" what tissue space the electrode is going through, gray matter vs white and also if a specific location based on parcellations?
I think this is clearly documented in the tutorials, please let me know if you think some part of the documentation is not clear.
https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Anatomical_labelling
For more information about this procedure, I recommend you refer to the section " 2. Electrode localization and anatomy" of this article:
https://www.sciencedirect.com/science/article/pii/S1053811922005559
Besides the table format of the information, how else can it be visualized?
The label of the current voxel in the selected atlas is displayed at the top-right corner in the MriViewer (but this is only one single voxel, not the most prevalent label in a sphere, as for the anatomic labelling menu).
https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#Subcortical_regions:_Volume
Any suggestion for the workflow since there are about 15 patients that I'm interested to analyze and each has about 10-16 electrodes totaling 200+ contacts and 5 recording sessions that are about 5 min each
In the first place, you could use a file format that documents correctly the channel names, so that you don't have to edit all the channels files.
If I edit channel file for one recording of 5min duration and there are 4 other recordings, can I copy the electrode positions and names from the first modified file to the others to save time instead of manually entering letters and repeating the registration/target/entry of electrodes?
If the acquisition context and the order of the channels is exactly the same: yes, you can copy-paste the channel file from the first file to the others of the same patient.
Moreover there are events that are marked in succession as a single timepoint at time at start of event and then the event ends when the next event starts, but the block of time is not marked from what I can tell.
I'm not sure I understand what this means. If you need help with events management, please post a create a new thread and provide some more context.
In the future I'm considering figuring out how to use mia to do group analysis of the different subjects recordings for trends of effects on SEEG signals from the events the amplifier generates/records. Seems like it may influence the answer to above questions.
@as_dub Do you have any suggestions?