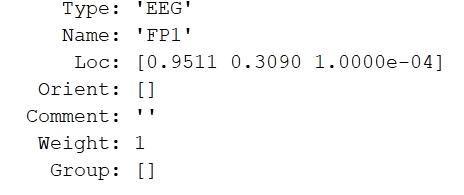
Hello everyone,
I am having trouble importing the EEG channel file from the BIOSEMI 128 system into Brainstorm. I had a spreadsheet of all electrode names and their coordinates, which I then transformed into a matlab structure through the following code:
% Get spreadsheet from Python
filename = "\BIOSEMI128.xlsx";
data = readtable(filename);
Channel = struct;
% channel array from table
for i = 1:128
Channel(i).Name = data{i, 1}{1};
Channel(i).Loc = data{i, 2:4}';
Channel(i).Type = 'EEG';
Channel(i).Weight = 1;
Channel(i).Orient = [];
end
% Headpoints structure
HeadPoints = struct;
HeadPoints.Loc = [];
HeadPoints.Label = [];
HeadPoints.Type = [];
% Finalize structure
chanFile.Channel = Channel;
chanFile.Comment = "BIOSEMI 128";
chanFile.HeadPoints = HeadPoints;
save("\channel_BIOSEMI128.mat", '-struct', 'chanFile');
To summarize, I just created the fields Channel (1x128 struct with fields Name, Loc, Type, Weight), Comment, and HeadPoints (empty 1x1 struct), forming them into a single structure saved into a .mat file. I based this off the structure of another .mat channel file of a different system that worked, but when I tried importing this one into Brainstorm, I got the following error:
Error: Line 667: No method 'add' with matching signature found for class 'org.brainstorm.tree.BstNode'.
**
** Call stack:
** >node_create_study.m>node_create_study/CreateNode at 667
** >node_create_study.m at 69
** >panel_protocols.m>CreateStudyNode at 577
** >panel_protocols.m at 46
** >tree_callbacks.m at 113
** >bst_call.m at 26
** >panel_protocols.m>CreatePanel/protocolTreeClicked_Callback at 156
** >bst_call.m at 28
** >panel_protocols.m>@(h,ev)bst_call(@protocolTreeClicked_Callback,h,ev) at 75
Could someone please help me out with this? Thank you!