Hello,
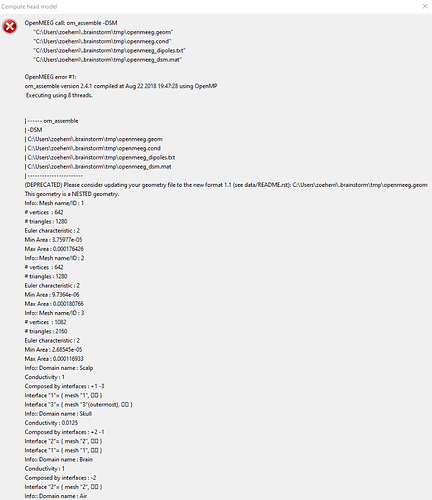
I computed a head model at the Cortex surface, then realized I needed a MRI Volume head model. I attempted to compute and was met with an error. I deleted the cortex head model and tried again but got the same error. I have attached the screenshot below. The message was so long I could not see the bottom. Do I need to download a different version of OpenMEEG? It automatically downloaded when I ran the first model.
Thank you!
@Alexandre @sik: any debugging suggestion?
@zoeherri
I think you already have the latest OpenMEEG version installed, no need to update anything.
Do you get this error also with different numbers of vertices in the BEM surfaces, or different patients?
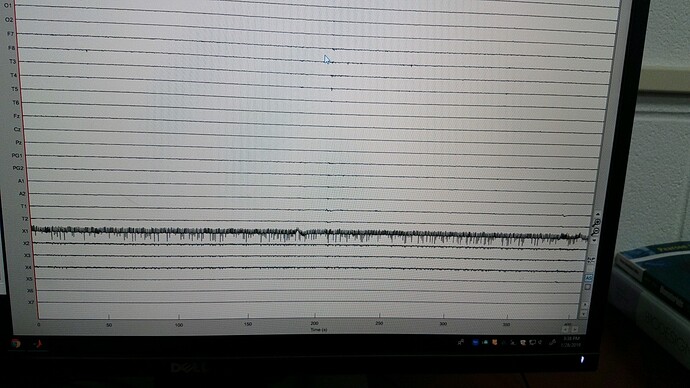
This is the first patient I have tried. We are using eeg data from a clinician that is imported as Nihon Khoden with 44 channels. The bottom 7 channels are labeled X1-X7 and are called file type DC. We are chatting with the clinician to determine what these are. There is one MISC type. The rest are EEG. We labeled the top 19 channels good, since these were clearly from a standard 10-20 system, and the rest were marked bad. We then right clicked the channels to add eeg positions, ICBM152>Generic>10-20 19, in order to compute the head model. I have attached a picture of one block of data with all channels on and can send more when I am in for the morning.
Thank you!
Right-click on the channel file > Edit channel file: Make sure all the channels you are not sure are EEG have a type that is not “EEG”.
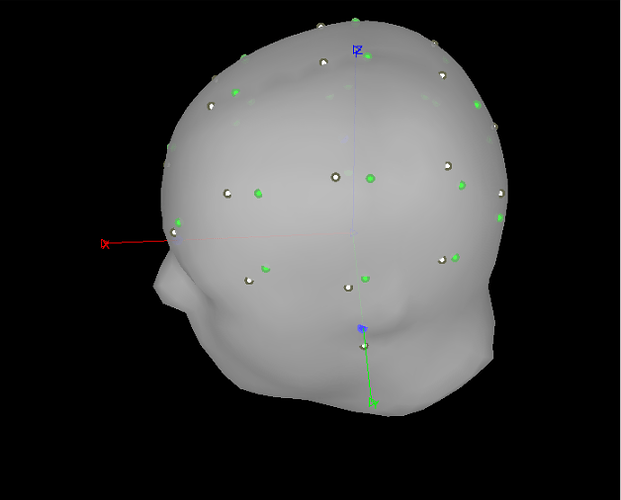
Then make sure all the anatomy looks good: double-click on the channel to get a view of the electrodes on the head, then add the BEM layers (in anatomy panel, simply double-click on them as well).
Please post a screen capture of the image you get.
@Francois With the exception of channel 20, which is MISC, channels 1-33 are all EEG. I added channels 21-33 back to the list of good channels. Then, I redid the EEG positions with ICBM152>Generic>ASM 10-20 94 and it pulled in 2 more EEG channels. The rest are called EEG_NO_LOC because they have names which do not coincide with the system. Is this what you meant?
All the channels that do not have 10-20 positions should be excluded from the head model computation. Having their type set to something else than “EEG” is the simplest way of excluding them (and “EEG_NO_LOC” works: it excludes them from the head model computation).
Are you using the MNI ICBM152 template or individual MRI scans?
Please post a screen capture of the 3D view showing electrodes + head, and another one with head + inner skull + outer skull + cortex.
If that is the case, then I have 21 channels with 10-20 positions and the rest are labeled as something besides EEG. Before I can compute the head model I need to right click the channel file and add EEG sensors. Was I doing this correctly previously?
We do not have MRI scans so we elected to use the default anatomy. I included a screen sketch of the MRI registration check as well. I have to put them in their own posts though because I am still a "new user."
I went back through the OpenMEEG tutorial and generated the BEM surfaces. I get a window that pops up after the window shown (image attached) that is not mentioned in the tutorial. Is this a newer addition to the program?
Once I generated the BEM surfaces I tried to compute the head model again. It took a lot longer to fail but ultimately gave me the same error.
@Francois Following the Volume source estimation tutorial, I ran it again but selected “Generate from cortex surface (adaptive)” and I got a head model. Why will it work for this option but not for the “Regular grid (isotropic)” option? We were hoping for this since our data is from seizures believed to be in the hippocampus and we would like greater resolution at these depth structures. Could it have to do with the estimated number of grid points due to the fine mesh I used in the last post?
Thank you for all the help here and with the tutorials.
Yes, you need to use the menu "Add EEG positions".
I went back through the OpenMEEG tutorial and generated the BEM surfaces. I get a window that pops up after the window shown (image attached) that is not mentioned in the tutorial. Is this a newer addition to the program?
No this is an option related to the fact that you are using a volume grid instead of a surface source grid.
See this tutorial: https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource
It took a lot longer to fail but ultimately gave me the same error.
I can see you are using a grid of sources with a resolution of 1mm. This produces a grid with 1.6 million points. The idea is good, but OpenMEEG is quite heavy in terms of memory and computation resources, and I bet this is not something your computer can handle. Try again with a 5mm grid and you will have a higher chance to get some results.
Francois
It worked when I used a wider grid. Thank you @Francois