Hello
I am working on functional connectivity development in children's brains using EEG. I have a question
To calculate the functional connectivity between brain networks, how can we adjust and select the scouts based on functional networks such as DMN, CEN, salience and etc.?
I would be grateful if you could help me
With respect,
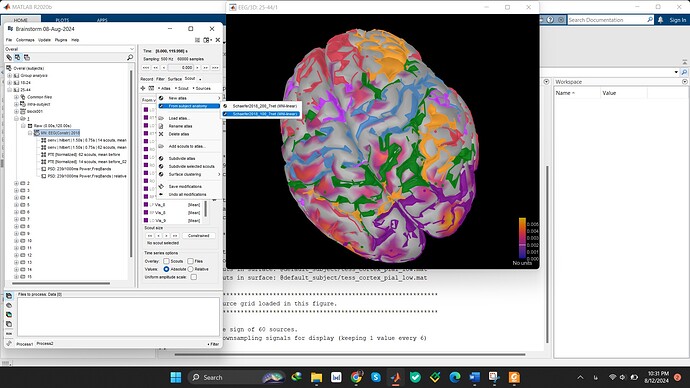
In Brainstorm, the Default anatomy includes the Schaefer (2017) parcellations. These parcellations are base on the brain network works by Yeo (2011). The Schaefer parcellations add further refinement by subparcellating the global networks based on a local gradient approach.
The Schaefer parcellations are available:
Relevant references:
thanks for your reply.
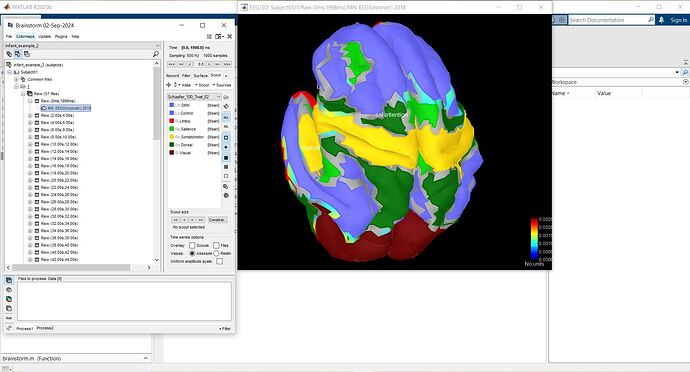
I tried this atlas and entered 100 parcellations, how can I also enter 7 network in scouts?
I would be grateful if you could help me
As you can see the cortical scouts derived from volume anatomy atlases do not work perfectly, those scouts are concentrated in the sulci, and gyri are not even part of scouts.
How was the MRI segmentation performed? Do you have access to cortical parcellations?
Hi
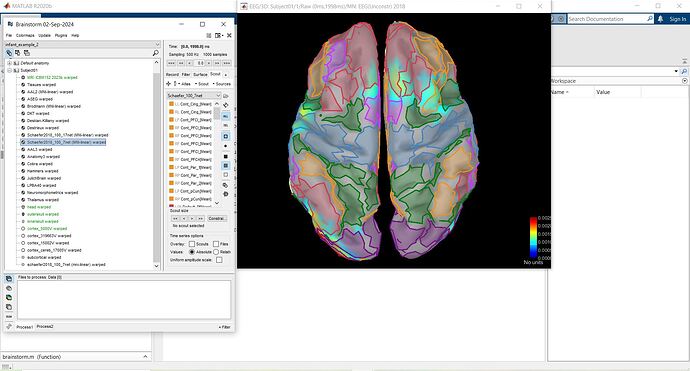
i am using wraped Default anatomy
do derived atlas (Schaefer 100 parcellations 7 networks) work correctly?
i have some questions
To calculate the functional connectivity diffirences between brain 7 networks can i use merge option to merge 100 parcellations ? calculation of the functional connection matrix can be based on the following figure, correct?
I would be grateful if you could help me
With respect,
Those are two different things, there may be confusion as the word atlas is used in different ways:
-
In the Anatomy view, the Schaefer2018_100_7net (MNI-linear) is an anatomical parcellation or anatomical atlas, i.e, a volume of integers where each value represents an anatomical label, and registered to an MNI space.
https://neuroimage.usc.edu/brainstorm/Tutorials/DefaultAnatomy#MNI_parcellations
-
On the Scout tab, the Schaefer_100_7net is a scout surface atlas, this is to say a collection of surface scouts, where each scout is a collection of vertices of the cortical surface. This surface atlas is computed during the MRI segmentation of the Default anatomy, so it is included in the warped anatomy.
https://neuroimage.usc.edu/brainstorm/Tutorials/DefaultAnatomy#FreeSurfer_templates
It sounds correct. Just be aware that when measuring connectivity between scouts implies dimension reduction (from source level to scout level), thus there are different ways to compute it, each with their advantages and disadvantages
https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence#Coherence_Scouts_x_Scouts