Hi dear Francois
thanks for answering my last questions.
i have questions about importing my anatomy folder, before i ask my questions, i wanted to say that i read the toturials number 1,2,3,4,5,6,7 and 20 and also EEG Epilepsy and i try myself with data tutorials step by step the tutorials and get the same results. then i decided to try it with myself data, because i wanted to create head model with my EEG data.
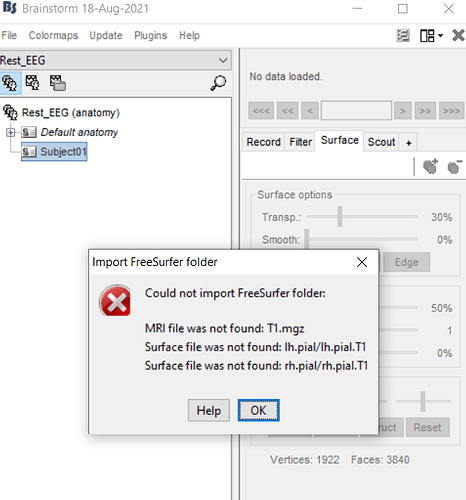
i instruct new protocol, and i make new subject,then i have been going to import anatomy folder but it makes error .
i wanted to ask you some questions:
1- for creating headmodels i nead MRI file and EEG data and Electrodes name and positions,is it true?
2-after importing anatomy and fuctional data is it necessary to preprocessing(averaging,bandpassfiltering,remove artifact) eeg data just for creating head model?
3-for EEG data and MRI and electrode position, is every format acceptable for Brainstorm?
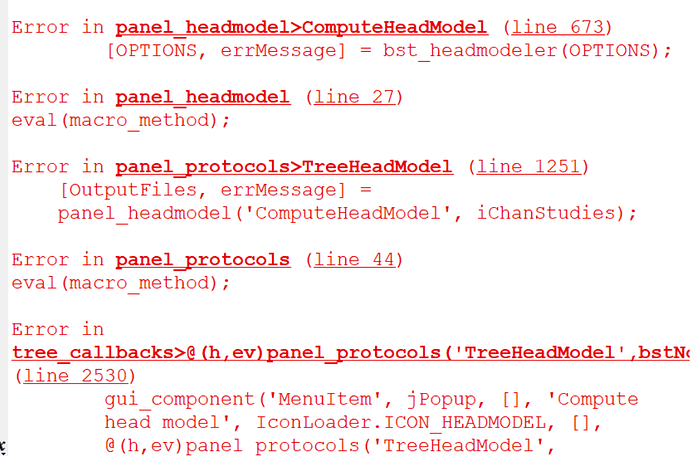
and my last question is why i encountered error when i import my anatomy folder?
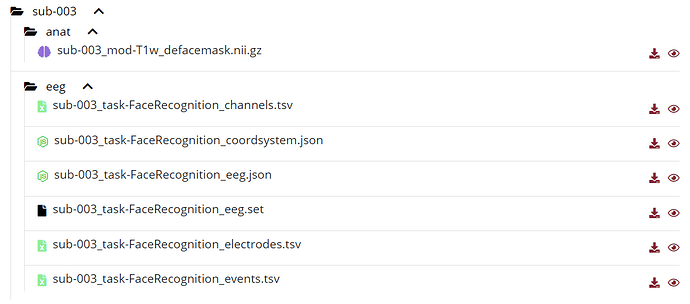
at the below i show you my folders format and my error comment
thanks so much
i have been going to import anatomy folder but it makes error .
If what you are trying to import is the BIDS folder sub-003/anat: it does not contain any FreeSurfer segmentation. Therefore it expected to give you the error you reported.
You need to run the MRI segmentation yourself, either from Brainstorm (with CAT12 for instance) or outside of Brainstorm (FreeSurfer, BrainSuite, FastSurfer...).
See the corresponding advanced tutorials:
https://neuroimage.usc.edu/brainstorm/Tutorials
If you want to import the MRI file from your BIDS dataset and then process it from Brainstorm, use the menu Import MRI instead of Import anatomy folder.
1- for creating headmodels i nead MRI file and EEG data and Electrodes name and positions,is it true?
Yes. As illustrated in the tutorials.
2-after importing anatomy and fuctional data is it necessary to preprocessing (averaging, bandpassfiltering, remove artifact) eeg data just for creating head model?
If you only want the head model to use it outside of Brainstorm: no, you don't need to preprocess/epoch/average the EEG recordings in Brainstorm.
If you want to estimate sources in Brainstorm: yes, clean/epoch/average your data first.
3-for EEG data and MRI and electrode position, is every format acceptable for Brainstorm?
http://neuroimage.usc.edu/brainstorm/Introduction#Supported_file_formats
Hi Francois,thanks for your answering my question...
as you siad, for importing MRI, i select the " import MRI " and then my MRI file is imported, then as you said, i should run the MRI segmentation, i wanted to do this with Brainstorm,so because of this i go to advanced tutorials: https://neuroimage.usc.edu/brainstorm/Tutorials.
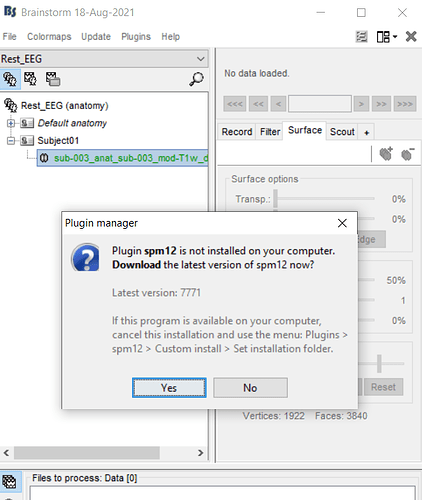
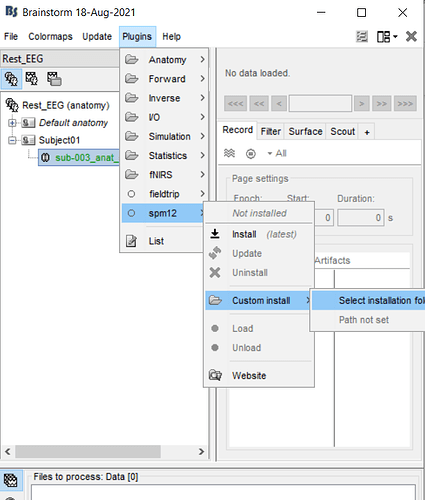
in this tutorial, at the part of "Run FreeSurfer from Brainstorm", we should go the path" Right-click > MRI segmentation > FreeSurfer ", but when i go to this path, there was not the option " FreeSurfer ", just there were the options: CAT12,spm12 ,...
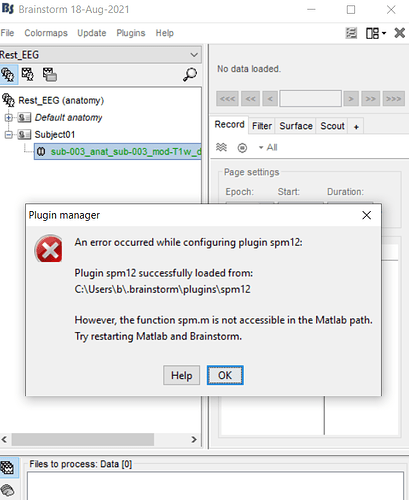
so at the next step i try with CAT12, then Brainstrom tell me that the spm12 should be plugins and i accept this, but then i encountered with error that i go to the link in my commandwindow and then i download the spm12, then open this zip file in the path of matlab, i do all of this steps, then comeback to Brainstorm toolbox and i encountered with the below error2
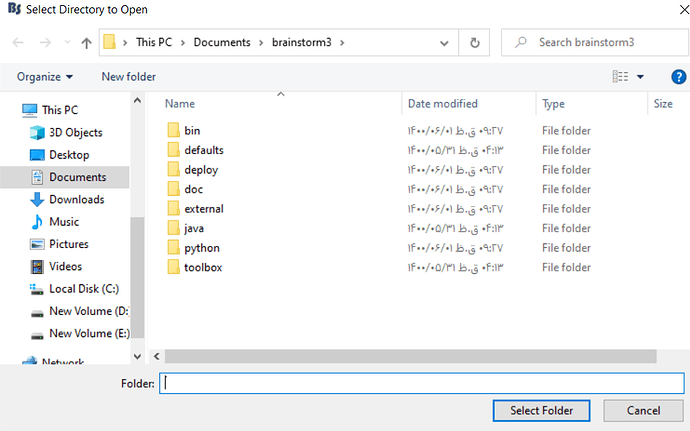
then i go to the plugins in the upper menu, and select custom install> set installation folder... then the window open, then i wanted to ask you, should i go to the matlab path and find my extracted spm12 files? and which file i should select now???
i mean that when i select the " set installation folder" and the window is opening... so what should i do? and which file i should select?
i am sorry that i ask simple questions from you because i am new user of Brainstorm and this is the first time i want to create head model with another EEGdata, not Brainstorm data, for my research field i should learn creating head model with this toolbox, and i need your help quickly... the time is very important for me ...
thanks so much ...
best regards
but when i go to this path, there was not the option " FreeSurfer "
so at the next step i try with CAT12, then Brainstrom tell me that the spm12 should be plugins and i accept this, but then i encountered with error that i go to the link in my commandwindow
Please be more specific on what this error is.
What is not working exactly?
then comeback to Brainstorm toolbox and i encountered with the below error2
What error?
Please copy-paste here the full error messages that you get.
thanks Francois
when i went to the option "plugins" in the menu, and i went the below step:
then the below window was opened for me:
and then i go to the derive C, the path where my matlab software in this path, and find the extracted file spm12 and select the folder "spm12" then i encountered with below error:
?????????????? help me please
thanks.
hi Francois
i answer your question... but you dont answer my question...
two days i waited for your answering, because for continuing my work, i need your answer...
Thank you for answering my questions
It looks like there is some confusion here: if you installed manually install SPM12, it should NOT be in the $HOME/.brainstorm folder.
Let's try debugging first the automatic install/management of SPM12, as this is what has the highest chances of working:
- Close Brainstorm
- Edit the Matlab path: remove all the folders except the Matlab installation folders
- Close Matlab
- Delete the folders
C:\Users\b\.brainstorm\plugins\spm12andC:\Users\b\.brainstorm\plugins\cat12 - Restart Matlab and Brainstorm
- Menu Plugins > spm12 > Install
- Menu Plugins > cat12 > Install
If anything goes wrong in one of these two last steps: please copy-paste the full error messages here.
hi
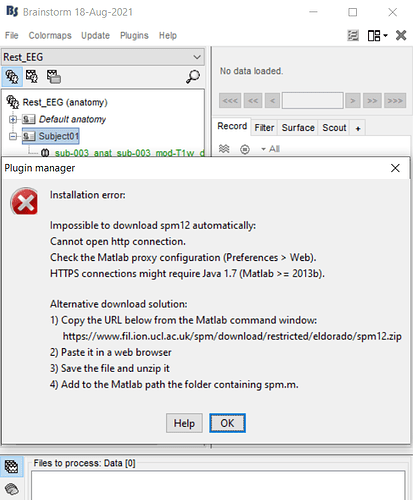
as you siad i do the steps, but when i wanted to install spm12, i encountered the below error:
and when i wanted to install cat12; the brainstorm tell me that i should install spm12 at first.
my matlab version is R2014b
Oh I see...
What version of Matlab are you using?
If you are behind a proxy, you could try configuring it in the Matlab options.
Can you get any file read with webread or urlread?
It would be worth investing some time to figure out how to download plugins...
Otherwise, if you want to try installing SPM12 and CAT12 manually:
- Close Brainstorm
- Edit the Matlab path: remove all the folders except the Matlab installation folders
- Close Matlab
- Delete the folders
C:\Users\b\.brainstorm\plugins\spm12andC:\Users\b\.brainstorm\plugins\cat12 - Download and install SPM12 not in any of the Brainstorm folders
Do not add the spm12 folder to the Matlab path. - Download CAT12 as described in the CAT12 doc: http://www.neuro.uni-jena.de/cat/index.html#DOWNLOAD
- Configure Brainstorm to use these custom installations for the two plugins, with the menu "Custom install": https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins#Example:_FieldTrip
hi
thanks for your last answers
my last problem about MRI segmentation has been solved...
but now for creating head model with my EEG data, i want to ask some question from you and your answer is important for me to have an accurate head modeling...
1- when i wanted to do MRI segmentation the Brainstorm ask me about some thing like thickness calculation or ... for computing and i should select "yes" or "no".., two small window opened and for the first window i select "no " and for the second window i select " yes ", and finally which created after segmentation is the below pic:
i dont have the item "cortex15002v " ...
2- when i read the tutorial "EEG Epilepsy", for solving forward problem the tutorial import event from file named "SPIKE", and then create head model for SPIKE, i want to ask you that for example for my EEG Epilepsy data ,not the Epilepsy Brainstorm data, should i have event or spike folder in my data folder, and should i import it ?? why ? the EEG data have the information about epilepsy so the separate folder named spike for what? and why when we want to create head model i should right click on the channel in the folder for spike in functional data?
why we shoud not use the eeg data?
3- in my data i dont have the name of the electrode, and i have just the number of them, for example 1,2,3,...,so can i use them? or i should have the name of them?
i mean that is it necessary to have the name of the electrode for exp(fp1,fp2,...)?
thank so much
Can you please describe how, for helping future users who will search the forum for help? - thanks.
1- when i wanted to do MRI segmentation the Brainstorm ask me about some thing like thickness calculation or ... for computing and i should select "yes" or "no"..,
What is your question?
This is probably documented here: https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12
2- when i read the tutorial "EEG Epilepsy", for solving forward problem the tutorial import event from file named "SPIKE", and then create head model for SPIKE, i want to ask you that for example for my EEG Epilepsy data ,not the Epilepsy Brainstorm data, should i have event or spike folder in my data folder, and should i import it ??
Please read carefully the tutorials.
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Mark_spikes
3- in my data i dont have the name of the electrode, and i have just the number of them, for example 1,2,3,...,so can i use them? or i should have the name of them?
i mean that is it necessary to have the name of the electrode for exp(fp1,fp2,...)?
This is up to you.
If you are expecting to use standard 10-10 positions, you need the names to match your data channels with the electrodes positions from the templates.
If you already have your electrode positions for each electrode, you can name them the way you want.
hi
thanks for your last answers.
i have some questions from you:
1- i try to create head model Using a large number of vertices, for example 1000000,but i cant, because when i select this number, and then i select the "MNI normalization" then i encountered below image:
my question is: why "Cortex 0v" ?
does it mean that Brainstorm cant have this number of vertice....?
for my research i should have a large number of vertices...the number "15000" is very small... so what should i do?
2- then i tried to create head model with the number of 15000 vertice, but with openMeeg model, the Brainstorm ask me that the OpenMeeg should install on your computer and i select "yes", but after 12 hours, the below window was still running:
and And it's not over yet, why?
3- And my last question is that when i created head model with my data, the item in the anatomy folder of the subject is:
i mean that the item ''cortex", and i have new item like "pial"? why? is "pial" equivalant with "cortex"?
thanks so much
You can't upsample the surfaces generated by CAT or FreeSurfer. The input number must be lower than the high-resolution surface.
I will add a test in the import function to make sure this cortex_0V file is not created.
for my research i should have a large number of vertices...the number "15000" is very small... so what should i do?
I really doubt that you can achieve anything interesting with a source space with more 300000 vertices, which you already have.
2- then i tried to create head model with the number of 15000 vertice, but with openMeeg model, the Brainstorm ask me that the OpenMeeg should install on your computer and i select "yes", but after 12 hours, the below window was still running:
Check the Matlab command window.
If there was an error: it would be displayed there.
Otherwise, it probably started the computation without updating the message in this window. If you selected the high-resolution cortex surface, it is plausible that it would take many hours to compute.
Open a resource monitor and check the memory and CPU usage.
If you have issues installing OpenMEEG automatically, see the instructions for custom installation of plugins: https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins
i mean that the item ''cortex", and i have new item like "pial"?
"pial" = "cortex" = external envelope of the cortex
"mid" = "central" = surface half-way between the pial and the white surfaces
Please refer to the introduction tutorials:
https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#Anatomy_folder
thanks
for installing openMeeg, i should say that the number of vertice 15000 the defult number, but more than 12 hours its continuing and then i close the window related to installing and cut it, i read the tutorial which you linked them for me, but it dose not solve my problem, if i wanted to install openMeeg manually, i have this link , but when i go to the section "download" i dont know which file i should download, can you help me?
or do you have better offer?
i should say that when i select installing openmeeg in my command window some error appear, look at below pic:
I'd recommend you try first to install OpenMEEG as a plugin.
Is it shown as installed in the plugins list?
Try uninstalling and re-installing it.
If you can't get this to work, try with 2.4.1 or 2.4.9999 for your OS.
i should say that when i select installing openmeeg in my command window some error appear, look at below pic:
- The error message is missing, please try again, then copy-paste the full error message (not a screen capture)
- This is not happening during the program installation
Hi
I have a question about head modelling.
I wanted to ask you that for calculating gain matrice, why EEG data is needed?
i think just MRI data and electrode positions and labels are needed...
and calculating gain matrice is just related to anatomical information of the head.
i mean that is it possible to achieve gain matrice just with MRI data and electrode positions and labels?
thanks
You don't need any EEG recordings for computing the forward model, you need only the anatomy and the electrode positions, indeed. Same thing for the inverse model:
https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Computing_sources_for_single_trials
hi dear Francois
i wanted to ask you about creating headmodel without electrode position, and just with MRI file and electrode label?
i have EEG data but i dont have its electrode positions. i just know that measuring EEG data was down in 10-20 system... is it posible to creating head model with this data with brainstorm?
how can i calculate the leadfield matrice for this data?
thanks alot
- Use the menu "Review raw file" to link the EEG recordings to your Brainstorm database
- Edit the channel file and make sure the names of the electrodes are standard 10-20 names (eg. Cz, FP7...)
- Right-click on the channel > Add EEG positions > ICBM152 > Generic >Select an entry that contains all your channels (10-20, 10-10 or 10-05)
- Adjust the template positions to the individual head (right-click on the channel file > MRI registration > Edit)
If there is any of these step that is not clear for you, follow the EEG tutorial first:
Tutorials/Epilepsy - Brainstorm
thanks
but when i link my EG recording with brainstorm and then i click on "Edit the channel file" some names of the electrodes is not in 10-20 system like (s1,s2,s3,s4,s5,s6,s7,s8,s9,s10,s11), now what should i do?
i know that the measuring EEG data in 10-20 system... ?????
accessing to these data leadfield matrice is important for me
thanks for your help