Then you need to ask the people who did the recordings the correspondence between these electrodes (s1,s2,...) and the 10-20 naming convention.
And then edit the channel file and rename these channels, before adding the template positions.
Or maybe they can provide you directly with a set of template 3D positions for this EEG cap. You could import these positions directly into your Brainstorm database.
unfortunately the people who record this eeg data dont information about these electrode because Because this data is taken from an article from 15 years ago and this data is a multi modal data about epileptic and the location of the lesion is specified by the doctor in the image file. i cant record EEG data with this property, in my country is not done for researchers, my research field is in brain localization and i should have a multi modal data like this data, so how can you help me? is there any other way?
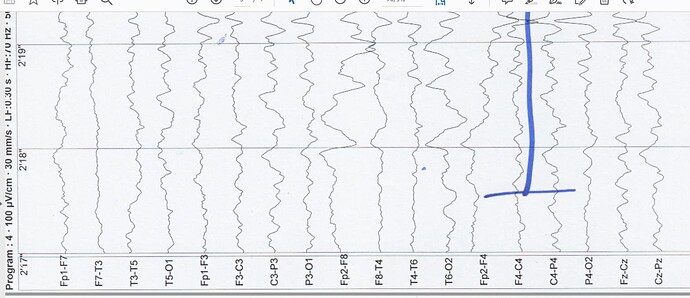
The only thing I have from this is the image below:
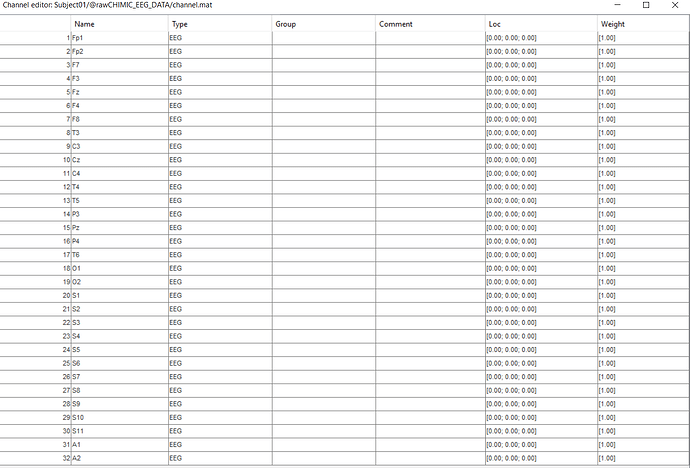
and then when i link this eeg data to brainstorm the electrode label are:
Could the electrodes S1-S11 and A1-A... be SEEG electrodes?
For the moment, change their types to something else than EEG, so that they do not interfere with the other channels. If you don't know what this is, there is no way you can use this information efficiently for source localization.
Make sure you follow completely the EEG/Epilepsy tutorial with the example database provided before trying to process your own recordings:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy
If you think this might be SEEG, you could be interested in these two other tutorials:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity
https://neuroimage.usc.edu/brainstorm/Tutorials/ECoG
hi
i want to ask you a question about the names of electrode in my data...
as i said before, the eleven electrodes which names are (s1,s2,...s10) are not in 10-20 system.
Can I ignore the data for these electrodes? And suppose I have only 20 electrodes in the system and consider their data?
as i said before to you, i dont have any information about the electrode position an i just have their names but some electrode names are not in 10-2- system, and i want to find their position with Brainstorm and then i create head model for this data, because of this i ask you above questions... thanks
Yes, edit the channel file and change their type to something else than "EEG", and they will be ignore when processing "EEG" channels.
Note that this is probably not enough to get any interesting source localization results.
hi
when we create headmodelling, we reach to locations of the surface elements(voxels) or gridloc, i want to ask you that can we have the location of the the center of the triangle on the surface? i mean that can we have the coordinates of the center of surface elemants not just the vertices?
thanks
When you have the vertices and the faces of a triangular mesh, you can compute easily the center of each triangle (mean of 3 vertices of each face) :
Centers = (Vertices(Faces(:,1),:) + Vertices(Faces(:,2),:) + Vertices(Faces(:,3),:)) / 3
Hi Froncois
i wanted to ask you about the coordinates of the Gridloc in MRIviewer
when i create headmodelling.i access to Gridlocation, and then i do source localization with my method in matlab and then i access to a voxel position which is the estimate of the source position, so i want to see this position in MRIviewer, but in which coordinates? we have four coordinates in it... with respect to below picture, i want to know that the position of the voxel in headmodelling in which coordinate?
thanks so much
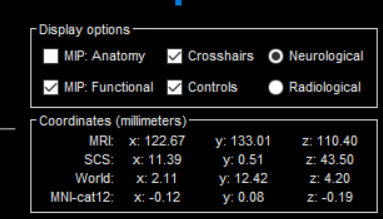
i want to know that the position of the voxel in headmodelling in which coordinate?
Most 3D coordinates in the Brainstorm database (channel positions, surfaces, source grids) are saved in SCS coordinates:
https://neuroimage.usc.edu/brainstorm/CoordinateSystems
For each new questions, please create a new topic, it makes it easier to search the forum for other users.
1 Like