Dear Brainstorm community,
does anyone of you tried to load the DISTAL atlas (Ewert et al., 2017) or a different deep brain atlas into Brainstorm and could me explain how to?
Greetings,
Matthias
Do you mean: other than what is described in this tutorial?
https://neuroimage.usc.edu/brainstorm/Tutorials/DeepAtlas
Dear Francois,
Thank you very much for the quick response.
Yes, it slightly different. I would like to include the DISTAL atlas since it is covering the Nucleus subthalamicus. As far as I now I can’t include this structure in the ASEG atlas provided by Freesurfer.
Greetings,
Matthias
If you have your atlas available as a volume (eg .nii file), you can import it using the menu “Import surfaces” and the file format “Volume mask or atlas”. You should be able to use this either in subject space or in MNI space. This would let you visualize the structures of interest as a surface.
https://neuroimage.usc.edu/brainstorm/News#Volumes_in_MNI_space
Otherwise, you can use the structures as ROIs with a volume source estimation:
https://neuroimage.usc.edu/brainstorm/News#Volume_atlases
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Volume_atlases
My apologies for the late response.
Since the Distal atlas is marking deep brain areas in the 2009b NLIN Asym ICBM space. I’m trying to follow your advice to include this atlas at first. I have the atlas as a .nii file. Surprisingly it takes a while to read it into Brainstorm using “Import surfaces” It is running already 5 days. Is this a possible duration or might there something be wrong with the file?
There is something wrong, indeed.
Have you figured out a solution to your problem?
Send me the files you are trying to import/process if you suspect there is any bug in Brainstorm
I’m still importing the atlas 2009b NLIN Asym ICBM. After ~12 days the import is done by 75%. The T1 file has a size of 135MB. Here you can find a download link: http://www.bic.mni.mcgill.ca/~vfonov/icbm/2009/mni_icbm152_nlin_asym_09b_nifti.zip
I will wait till the import is done. So I can see if there might be also a problem with the imported data.
If there might be a problem with the DISTAL atlas i just can see after the first import is done.
The import stopped with a warning: “Index exceeds matrix dimensions”
And so far I couldn’t find the atlas in the database.
Since I haven’t mentioned it before I used the option “Import surfaces” to import the atlas.
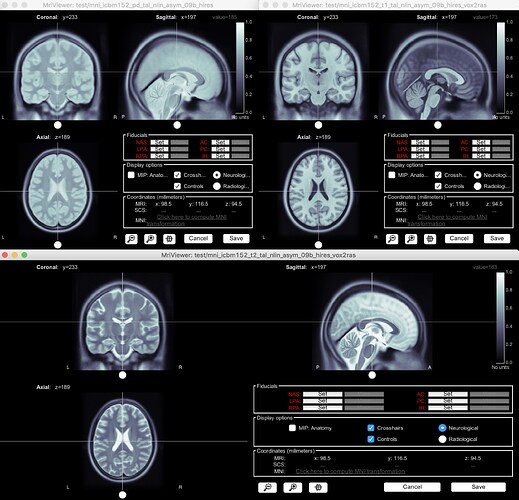
Hello:
I was able to import all 3 volumes: int was quick. See screenshots. I ignored the reslicing/reorientation prompts.
Make sure you have selected Nifti-1 from the file format pulldown menu after you have selected: Import MRI volume (not folder) from the subject database.
Hope this helps
Matthias:
I’m not sure I understand what you are trying to do… If you want to use the ICBM152 template: just use the default anatomy in Brainstorm, available in all the protocols.
I was suggesting you use the menu “Import surface” only for loading your atlas, but now I’m not sure this is a good recommendation for you.
Can you describe precisely what you are expecting from Brainstorm regarding this DISTAL atlas?
Hi,
the DISTAL atlas is including information about the STN and other subcortical structures.
I have LFP’s measured in the STN and i would like to register a individual MRI with an template which contains subcortical structures especially the STN. Based on this information I would like to calculate a leadfield. Basically I want to do something like described in the DBA tutorial, just including the STN. And for the beginning I wanted to use a template MRI. But so far I’m failing including the STN. If I check the position in the MRI viewer, it is obviously displaced.
I was referring to the atlas 2009b NLIN Asym ICBM since in LEAD DBS they are using it as a base for the DISTAL atlas. I wasn’t aware that it was just a subversion of the ICBM152.
The DBA tutorial is based on some research work from Attal et al., which includes only the brain regions listed in this tutorial. You won’t be able to compute any accurate forward model from additional meshes you would import manually.
What you could do instead is to compute a volume forward model, then load your atlas as regions of interest:
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Volume_atlases
If this is only for display purpose, you should be able to use the menu “Import surfaces” with the file format “Volume mask or atlas (MNI space)”.
If one or the other seem to create ROIs that are out of place, please send me the atlas volume you are trying to load, I’ll try to understand what is wrong with it.
Thank you very much for all the help.
Both ways are working fine. Also the ROI’s are placed correctly.