Hello everyone, we are currently working on a project that involves analyzing MEG data using Brainstorm. We have generated a scout (region of interest) as a .mat file with vertices data, and we also have ICBM152-based whole brain vertices and faces. Our next step is to generate a mask as a NIFTI file of our scout, which we plan to use for whole brain spectroscopy analysis with MINT/MIDAS. However, we've encountered some difficulties with this process. We've tried using the vertices in the .mat file from ICBM152_aseg to locate the scout in an ICBM152 template, but the vertices do not seem to locate the scout as we see in Brainstorm. We're wondering if anyone has any insights or suggestions on how to apply these vertices and use the .mat files to generate the NIFTI file we need. Thank you for your help!
To export scouts as .nii files, I would recommend using a volume source model instead. A volume scout can be directly exported as a .nii file from the Scout tab, menu Scout > Export as MRI mask.
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource
Exporting a surface ROI as a volume mask is not a trivial problem, that we decided not to handle from the Brainstorm interface.
If you really want to do this, you can try to reuse some of the code that was developed for the menu "Scout > Edit in MRI", which we don't recommend using anymore as its behavior is difficult to understand. Function EditScoutMri() in file panel_scout.m.
Hi Francois,
Thank you for your suggestion on exporting scouts as .nii files using a volume source model. I will try that out. However, I still have a question about using the function EditScoutMri() in file panel_scout.m, as you suggested. I have tried to rename this file as process_panel_scout.m and put it into the "$HOME/.brainstorm/process" folder, but it doesn't seem to be working. Could you provide some guidance on how to properly use this function? Additionally, we are still experiencing difficulties using the vertices in the .mat file from ICBM152_aseg to locate the scout in an ICBM152 template. Do you have any insights on why this might be happening or any suggestions on how to address this issue? Thank you for your help.
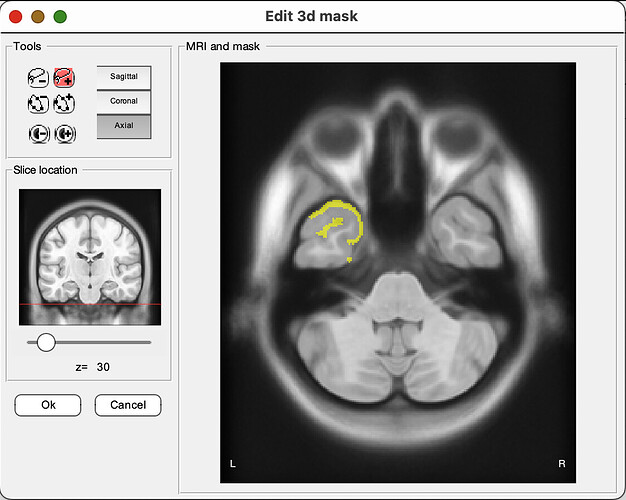
For example, we would like to export the following mask as a NIFTI file:
However, I still have a question about using the function EditScoutMri() in file panel_scout.m, as you suggested.
I did not suggest to use this function, I pointed at some code that could be useful for a Matlab developer to reuse, in order to create new tools, that are not available at this moment.
If you want to draw volume masks, I'd recommend you use tools that are properly designed for that (MRIcron, 3DSlicer...)
I have tried to rename this file as process_panel_scout.m and put it into the "$HOME/.brainstorm/process" folder, but it doesn't seem to be working. Could you provide some guidance on how to properly use this function?
Process functions are plugins with a specific design. You can't rename any function and expect it to work as a plugin. See the documentation for developing processes:
https://neuroimage.usc.edu/brainstorm/Tutorials/TutUserProcess
Additionally, we are still experiencing difficulties using the vertices in the .mat file from ICBM152_aseg to locate the scout in an ICBM152 template.
I'm sorry, I don't understand what you are trying to do here.
Search the website forum for help with using scouts and surfaces programmatically.
https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_Surface
https://neuroimage.usc.edu/brainstorm/Tutorials/Scouts#On_the_hard_drive
https://neuroimage.usc.edu/brainstorm/Tutorials/Scouts#Additional_documentation
Hi Francois,
We were hoping you could guide us towards someone who may know why our BRAINSTORM-generated scout (a Matlab file containing vertices with their corresponding x, y, and z spatial coordinates from the ICBM 152 atlas) does not correctly map the scout's ROI in an ICBM 152 atlas using SPM 12. We are confident that the Matlab file somehow represents the ROI (all vertices cluster together inside the brain), but we need to find a transformation to shift these vertices to what we can see in BRAINSTORM as the real ROI.
We would really appreciate any pointers you can give us
The scouts definition in Brainstorm contain only indices, not coordinates.
From the cortex surface file, you can get the coordinates in SCS coordinates.
If you want the coordinates in MNI coordinates or in WORLD coordinates of the ICBM152 .nii file, you need to do this conversion explicitly.
All the information about the various coordinate systems and their conversion is available on this page:
https://neuroimage.usc.edu/brainstorm/CoordinateSystems
Can you please post here a clear example of what you are doing?
Including screen capture of everything you are doing step-by-step, and screen captures illustrating why you are not satisfied with the results you get.