Hey Yvonne,
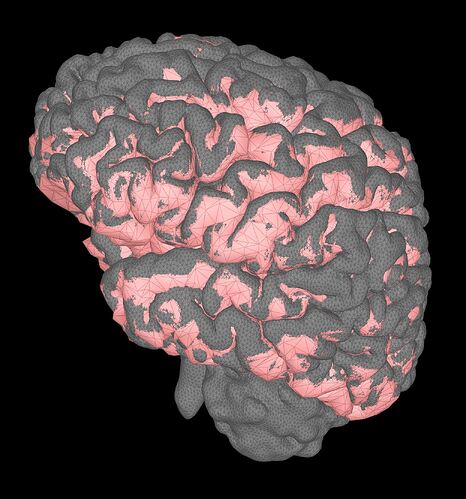
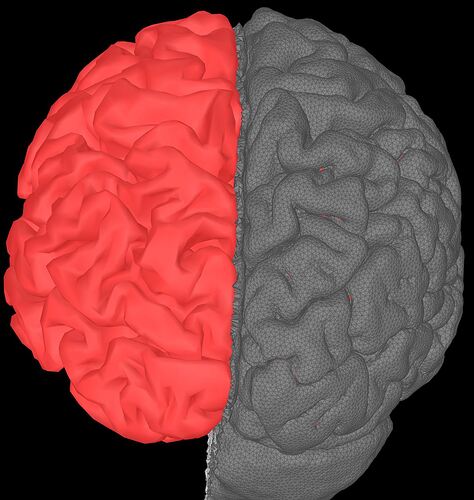
Visually, the fast way is to display the cortex and the mesh, and then check if the cortex is totally inside the GM volume, similar process with the WM
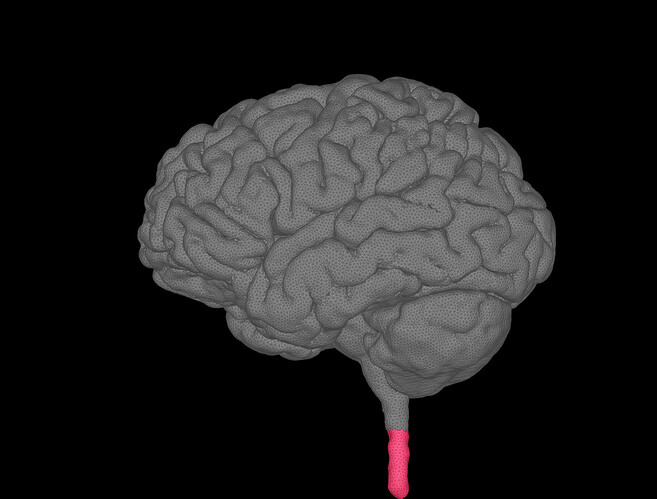
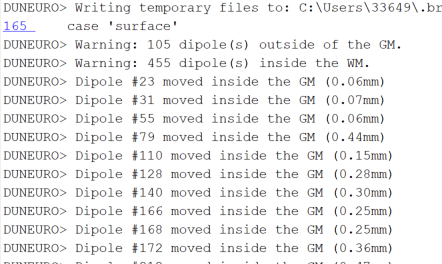
Here is an example of a bad dipole location:
in red is the cortex surface (dipole locations) and in gray the surface of the GM.
The requirement for the FEM (also for the other method) is to have the neural source within the GM. FEM is more complex since there are different source models.
If you don't there is a high chance that the solution is not correct.
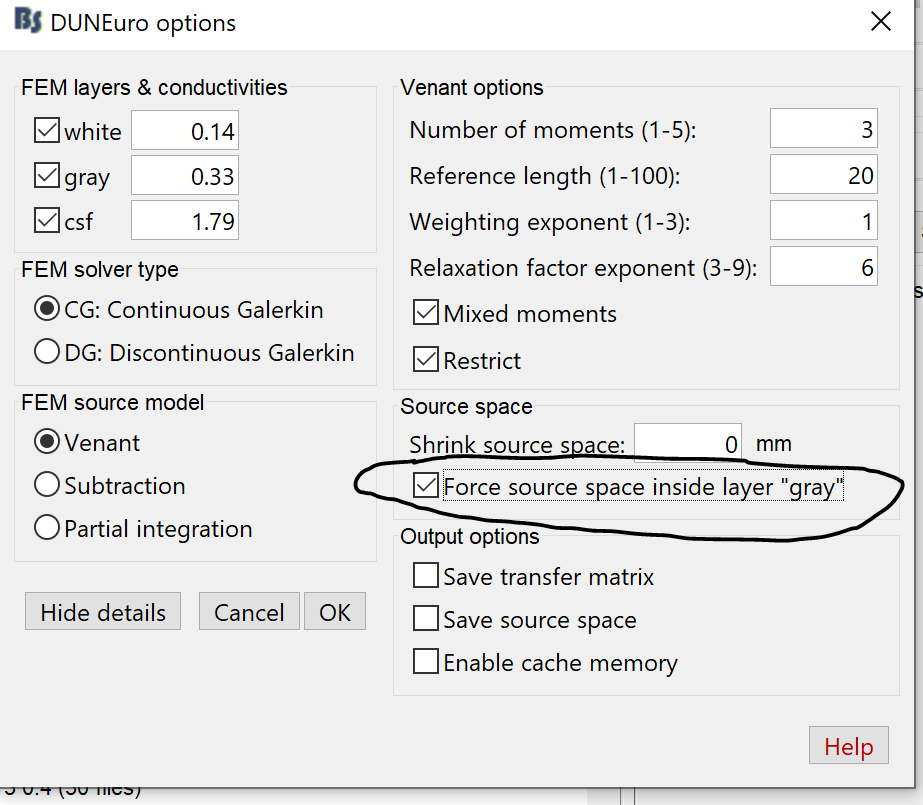
Practically, to prevent this problem, you need to use the option "force inside ... ", with this, all the dipole outside the GM are moved slightly to their nearest centroids of the GM element and ensured to be within the GM
The other solution, is highlighted by the second option "Shrink source space" by xx mm, in this case, the cortex surface is "deflated?" or shrined by xx mm inward following the normal direction for each vertex.
You can use both these options together and then save the FEM source space if you want to check it.
indeed, you are right, we are checking this issue with Christian/Carsten.
Right now, we think that a random initialization of the process is applied and then it can work in some cases ... if you are lucky 
We are going to fix this asap.
The other solution is to mesh the air bubbles inside the skull that are the reason for this error.
This is weird, which version of Brainstorm do you have (date of your last update)?
What is the size of the cortex and the FEM mesh that you are using?
Maybe I need to reproduce this error with your data.
Let me know, otherwise, I need to check it with your dataset 
Best,
Takfarinas