Hello,
So I'm working on trying to analyze some data using Brainstorm.
I'm using the non-Matlab version of Brainstorm since
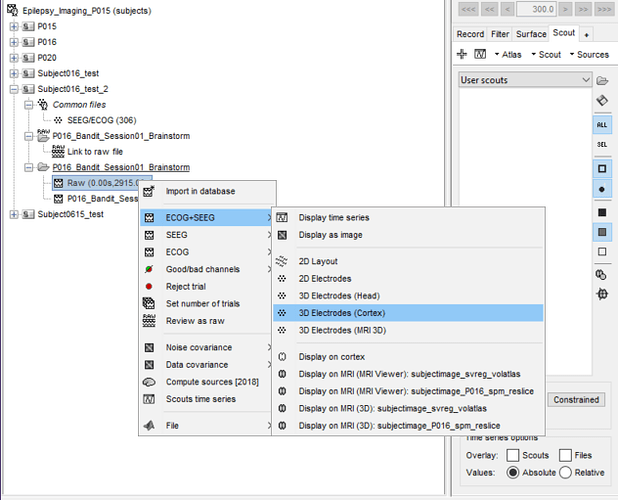
I've loaded a file into brainstorm and this is what the system is showing right now:
I'm right-clicking on the raw file, going to SEEG+ECOG, and then I'm clicking on display on cortex.
However, when I click on "display on cortex", my computer makes the windows error noise and I get this loading:
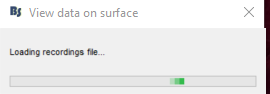
The file never loads and doesn't show up on the cortex.
What am I doing wrong?
Thanks!
Sincerely,
Arul
In the terminal window that is opened when you start Brainstorm, you should be able to see an error message.
Could you please make a screen capture of this error message and post it here?
Thanks
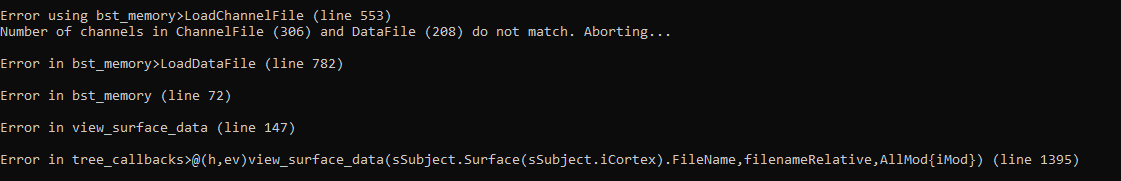
This is the error message I'm getting. From what I'm seeing, the channel fild and the data file don't have the same number of channels. However, according to the other members of the lab that I'm working in, this patient did have 306 electrodes. What could be going wrong?
Thanks!
Sincerely,
Arul
This is most likely a problem with the way you imported and processed your data.
- When creating a new protocol or subject, avoid using the option "Default channel file: Yes", this is less safe when you are not really confident with what your are doing in terms of data structures
- Never import a create a channel file from nothing: Always start from linking the recordings to the database, and then use the menus
Add EEG positions or MRI registration > Edit.
- Before processing your own recordings, start by following all the tutorials dedicated to EEG/SEEG/ECOG data:
Hello Francois,
Thank you for your speedy response! I created a new subject and first added the recordings and this time it worked!
Sincerely,
Arul