Hi, I want to import some ASCII EEG resting-state data into Brainstorm. This is the first time I use Brainstorm for EEG and for resting-state data.
The aim is to produce the source reconstructions and then get the time series of the scouts based on the Yeo atlas at the individual level.
So, I have created the database by importing the functional data (EEG 57 channels). The preprocessing (filtering, ICA) has been performed some time ago in Fieldtrip so I have just imported 15 segments of 4 sec each per participant as ASCII text files.
I have performed all the steps on two subjects to get to the scouts based on Yeo, but I have some questions and also need some guidance to improve the channel file.
Anatomy
I don't have the individual MRIs so I considered using either ICBM152 template anatomy or the FSaverage. It is mentioned in the tutorial 'Using the anatomy templates' that 'all templates include similar information but Yeo2011 and PALS are for FSAverage only'. It also includes Brainnetome which I plan to use for another ERP analysis. However, I can't say that I understand this distinction.
Q1. Why can't ICBM152 contain these parcellations? This interests me mainly because as you will see later on the channel file is registred correctly on the ICBM152 but needs some modification for Fsaverage.
Q2. Also, I did not understand what is the use of the atlases (e.g. Brainnetome, or Schaefer atlas) if I download them in the anatomy folder? Is it only for visualization purposes? Is there something that I should do at this point to merge/project the atlas on the template anatomy, or is this the reason to use Fsaverage in the first place? At a later stage, when choosing the scouts I realized that I don't need these atlases in the anatomy folder, but still, I am not sure about their use.
Channel file
I have two versions of the channel file, one created in BESA (elp, with Phi, Theta values) and an xyz file (Cartesian values) (both attached). The cap is from easyCap 64 channels. I have read that either XYZ Theta and Phi are real 3D positions, they are all spherical, so they are not of much use if the aim is to perform source solutions.
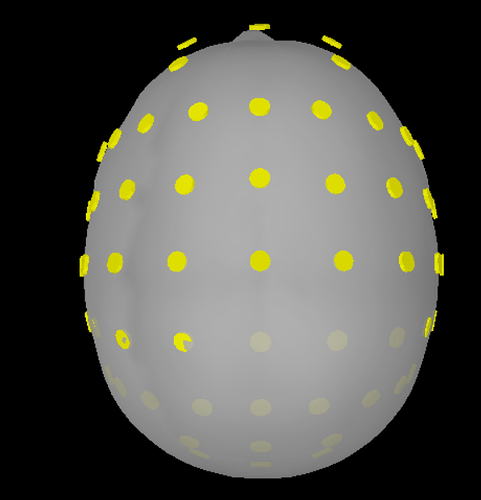
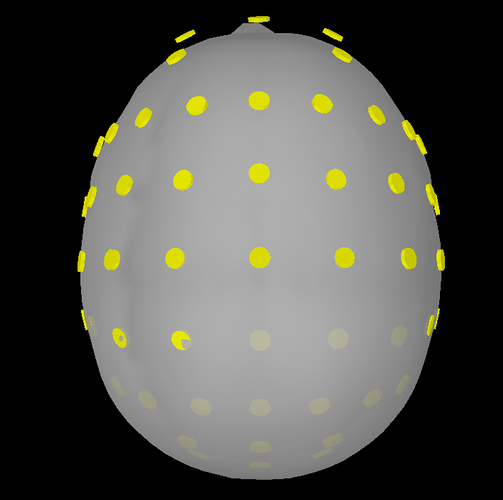
I have tried to import the elp file as a BESA file but it needs some modification. Typically the BESA elp has 4 columns: the first one is the type of sensors (e.g. EEG, POL, etc), then the label, and finally Theta, and Phi values. You simply have to remove the first column. Then, I used the command 'import channel file' and accepted the scaling factor of 0.1 and the MRI transformation. but still, the sensors are not placed correctly on the head (please see BESA 1.png). I've added the electrode positions based on the Brain Products Easycap 64, but this resulted in losing one sensor, the AFz and generally having many electrodes dislocated (please see BESA 2.png). Here, the anatomy is based on the Fsaverage template and the result needs a lot of work. When choosing ICBM as the template, the result is not so bad and only 1-2 sensors would require some manual adjustment (though the AFz would still have to be introduced from the beginning).
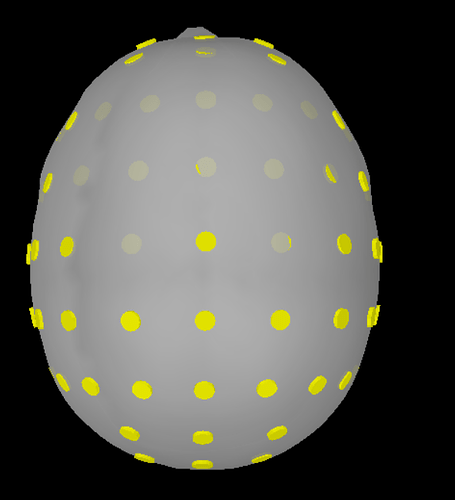
Alternatively, I've imported the XYZ file (there is some (slight) difference when choosing XYZ, Name vs XYZ_MNI, Name), and accepted the scaling factor of 0.1 and the MRI transformation. The end result needs modification probably on all sensors (please see XYZ 1.png) but it seems better from the result based on the elp file given that AFz is preserved and that the electrodes seem less displaced (but still displaced). I did not use the command 'add EEG positions' as this results in almost the same registration as with BESA but with AFz preserved (please see XYZ 2.png). If ICBM152 would be the desired template minor modifications would be required (please see XYZ 3.png)
***Q3.***Is the only way to tackle this issue to manually modify the XYZ 1as proposed in another post? Is this based only on visual inspection of the result? I am worried that the end result may not be accurate enough.
channels.elp (1023 Bytes) channels.sxyz (1.3 KB)
BESA 1.png
BESA 2.png
XYZ 1.png
XYZ 2.png
**Q4.**Why is there so much difference in the registration of the electrodes on the MRI when the template is ICBM152 vs Fsaverage?
XYZ 3.png
Noise covariance
Reading the tutorials, I've decided to go with Option #1 for EEG in resting-state, that is, calculate noise covariance over a long segment of resting recordings, but save only the diagonal elements, i.e. the variance measured at each sensor. Option #2: Select "No noise modeling" assumes that noise is equivalent on all sensors, by this dataset was a bit noisy.
Q5. In this case, should the baseline time window be the same as the data window?
Q6. Does selecting all 15 segments of 4s per subject and computing the noise covariance based on these (total 1 minute), satisfy Option #1? How long should the resting state segment be at a minimum? If shorter than the minimum, then chose Option #2, or is there something else to take into account?
Q7. A more theoretical question. I often submerge the EEG electrodes in saltwater to check if a sensor should be replaced. Could such a measurement (with electrodes submerged) prior to the actual recording be something equivalent to empty-room measurements for MEG and thus be used for noise covariance estimation?
Thanks in advance,
Best,
Haris