Dear BST Community,
I am having trouble using the Envelope Correlation N x N (2020) feature.
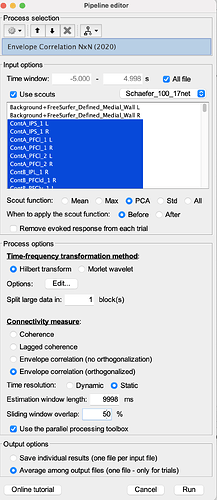
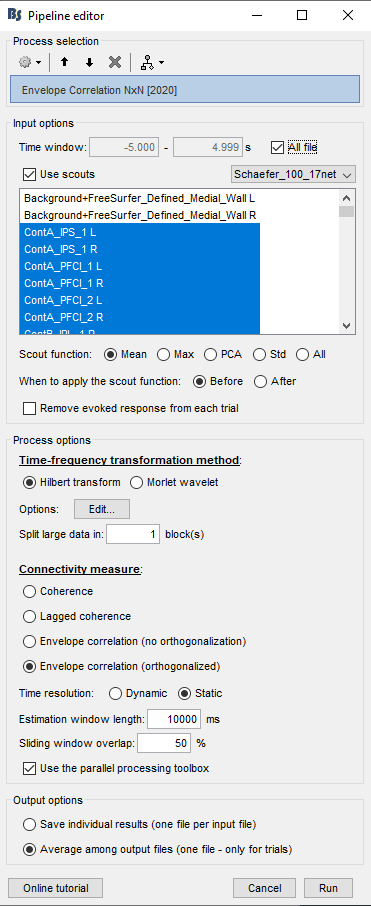
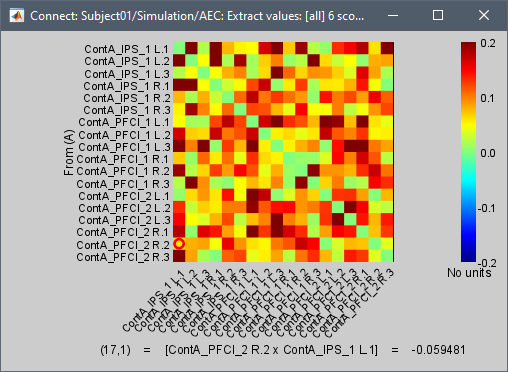
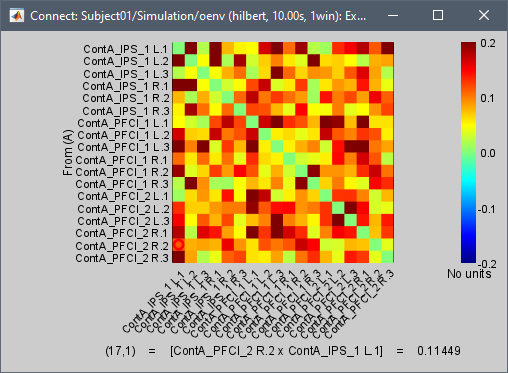
I am testing it on one participant, with 47 epochs (source localized), each epoch of 10 seconds long (512 Hz samples). Running Amplitude Envelope Correlation (N x N) works and provides no error, but to speed up the process, I would like to run the exact same analysis but with the parallel processing provided in the Envelope Correlation N x N (2020).
My understanding is that the "Estimation window length" should equal to the "Time Window" if I want to produce the same results as the Amplitude Envelope Correlation (N x N).
But when trying to do so, I am running into the following error:
** Error: [process_henv1n] Connectivity > Envelope Correlation NxN (2020)
** Line 23: cat
** Dimensions of arrays being concatenated are not consistent.
**
** Call stack:
** >process_henv1n.m at 23
** >bst_process.m>Run at 230
** >bst_process.m at 37
** >panel_process1.m>RunProcess at 124
** >panel_process1.m at 26
** >gui_brainstorm.m>CreateWindow/ProcessRun_Callback at 768
** >bst_call.m at 28
** >gui_brainstorm.m>@(h,ev)bst_call(@ProcessRun_Callback) at 292
**
**
Below is my GUI setup:
Any help with this would be greatly appreciated.