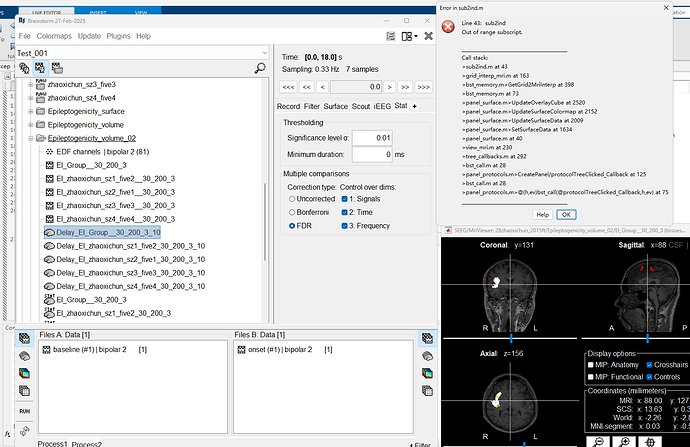
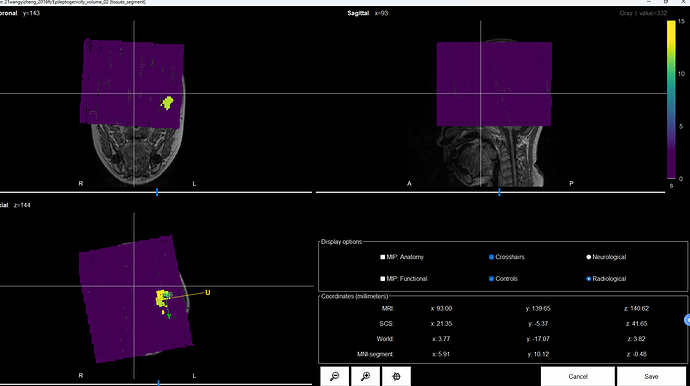
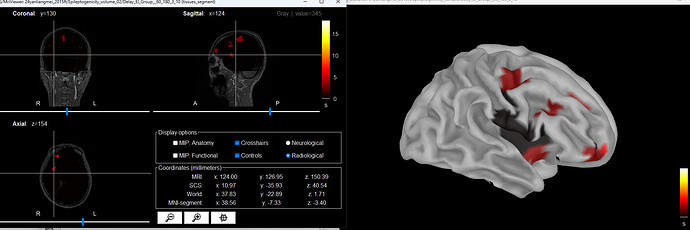
Hi, I encountered an error while generating the delay map for the volume map. The error in the command window is as follows:
** Error: Line 43: sub2ind ** Out of range subscript. **
** Call stack:
** >sub2ind.m at 43
** >grid_interp_mri.m at 163
** >bst_memory.m>GetGrid2MriInterp at 398
** >bst_memory.m at 73
** >panel_surface.m>UpdateOverlayCube at 2520
** >panel_surface.m>UpdateSurfaceColormap at 2152
** >panel_surface.m>UpdateSurfaceData at 2009
** >panel_surface.m>SetSurfaceData at 1634
** >panel_surface.m at 40
** >view_mri.m at 230
** >tree_callbacks.m at 292
** >bst_call.m at 28
** >panel_protocols.m>CreatePanel/protocolTreeClicked_Callback at 125
** >bst_call.m at 28
** >panel_protocols.m>@(h,ev)bst_call(@protocolTreeClicked_Callback,h,ev) at 75
The error seems to be caused by the t-value interpolation exceeding the boundary. What should I do?
The error does not seem related to the t-value themselves, but to the interpolation between the masking of the default surface to the MRI volume.
Could you provide more details? or steps to reproduce the error?
- Are you using the Default anatomy?
- Can you share a screenshot of the Anatomy files for the Subject?
P.S. Please do not repost a question to put in on top of the board. We answer all the posts as fast as we can. The other thread with the same question is now deleted.
I sincerely apologize for not following the forum posting rules, which has caused additional workload for you and your team. There is no issue loading the Epileptogenicity map in volume with no delay, but there is an error when trying to load the delay map in volume.Below is the error message from MATLAB.
** Error: Line 43: sub2ind
** Out of range subscript.
**
** Call stack:
** >sub2ind.m at 43
** >grid_interp_mri.m at 163
** >bst_memory.m>GetGrid2MriInterp at 398
** >bst_memory.m at 73
** >panel_surface.m>UpdateOverlayCube at 2520
** >panel_surface.m>UpdateSurfaceColormap at 2152
** >panel_surface.m>UpdateSurfaceData at 2009
** >panel_surface.m>SetSurfaceData at 1634
** >panel_surface.m at 40
** >view_mri.m at 230
** >tree_callbacks.m at 292
** >bst_call.m at 28
** >panel_protocols.m>CreatePanel/protocolTreeClicked_Callback at 125
** >bst_call.m at 28
** >panel_protocols.m>@(h,ev)bst_call(
@protocolTreeClicked_Callback,h,ev) at 75
**
Here is the Anatomy files of the Subject
 This issue was mysteriously resolved after I modified the electrode spacing, but the interpolation results look a bit different from the normal ones. Why is that?"
This issue was mysteriously resolved after I modified the electrode spacing, but the interpolation results look a bit different from the normal ones. Why is that?"
In the shared anatomy, there is no surface (nor cortex, nor skull), that is used to compute the brain mask, thus the source grid looks strange, it comprises places outside of the head.
Thank you very much for your guidance. The new issue is that at time = 0, the color of the seizure onset region seems to be the same as the surrounding non-relevant voxels, which makes it difficult to locate the onset region in the volume. Is there any way to distinguish them?
Just change the colormap to something where zero is not black
https://neuroimage.usc.edu/brainstorm/Tutorials/Colormaps