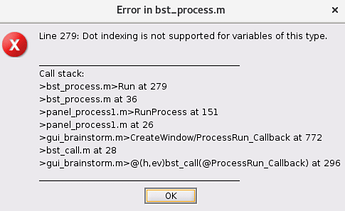
Dear Brainstorm team,
Whenever I try to perform any transformations on scouts (i.e., Hilbert, FFT, PSD etc) I get the following error -
I have taken the following steps before arriving at this error -
- Preprocessed data were imported in database
- Source analysis was performed (dspm)
- Data were projected on standard template
- Spatial smoothing (7mm FWHM, not rectified)
- Save some ROIs from Desikan-Killiany atlas - import them as user scouts
- Drop the source file (from group analysis) into process1
- Frequency -> Hilbert Transform -> "Use scouts (select appropriate ones)"
- Run
The process stops with the error (see above). Turns out it does produce the output (Hilbert TF plot) but I only see it after I have restarted Brainstorm.
Please let me know if you have any suggestions.
Thanks
P.S. I am on the latest version of bst, running in Matlab 2018a. OS: CentOS Linux 7 (64 bit)
I could not reproduce the error you reported.
I added a test to avoid seeing the error you reported, but it will not make it work either, the output file would not appear...
https://github.com/brainstorm-tools/brainstorm3/commit/23eb266a3116516358ae6523c448e5f89f637d79
Something might be wrong with the structure of your database. Don't you see any other warning messages in the Brainstorm command window?
Go to the anatomy view, right-click on the top element in the database explorer > Reload.
If you still get an error, please post a screen capture showing the input (projected sources) and output files (time-frequency).
About your processing pipeline:
- If you are interested in the TF decomposition of a scout, do not project the sources to the anatomy template first: compute it directly from the subject results.
- You don't need to copy the DK scouts to the "User scouts" at;as
- Smoothing min norm sources without rectifying the signals is not advised, it may end up averaging nearby positive and negative values.
Some reference pipelines are described in this tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/Workflows#Constrained_cortical_sources
Hi Francois,
Thanks for the prompt reply.
The test that you have added is certainly helpful as I have noticed something strange.
For both the "group analysis" files and individual subject files, I get the same error message:
BST> Warning: 1 file(s) not found in database.
I have followed your suggestion of reloading the files as well and the error still persists.
The contents of both the files are here -
- Subject file
- Projected and spatially smoothed (same subject, same epoch)
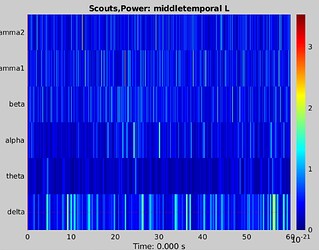
- TF plot after restarting bst
The data are from a continuous listening experiment so there are no ERP components per se but this does look a bit odd (perhaps its just this subject). Our aim is to use the PAC (and tPAC) tools in bst. I couldn't find any documentation on group analysis using PAC (happy to contribute that if I get this work).
My suspicion is that this error has occurred when the files were written so I probably will have to re-compute dspm. What do you think?
So the revised pipeline will have to be -
- preprocess data
- Import epochs into database
- Compute dspm
- Use ROIs to compute PAC
Key question here is how do we know that we are comparing the same ROI between participants (i.e., due to differences in individual anatomy. Some participants have individual MRI, some don't)?
Apologies for writing an extensive reply but I really do appreciate your help.
Cheers
Then there is indeed some error in the structure of your database.
It might be an error that is not even related with these files.
Maybe duplicate your protocol and delete files until you find where the error is.
The data are from a continuous listening experiment so there are no ERP components per se but this does look a bit odd (perhaps its just this subject)
What looks odd to you?
Our aim is to use the PAC (and tPAC) tools in bst. I couldn't find any documentation on group analysis using PAC (happy to contribute that if I get this work).
@Samiee @Sylvain?
My suspicion is that this error has occurred when the files were written so I probably will have to re-compute dspm. What do you think?
I can't guess what went wrong in your previous manipulations...
Make a backup of everything before it gets worse, and try to identify where the error is coming from.
Key question here is how do we know that we are comparing the same ROI between participants (i.e., due to differences in individual anatomy.
If you are using an anatomical atlas generated by FreeSurfer, you know that a given ROI represents the same anatomical structure across subjects.