Dear Francois,
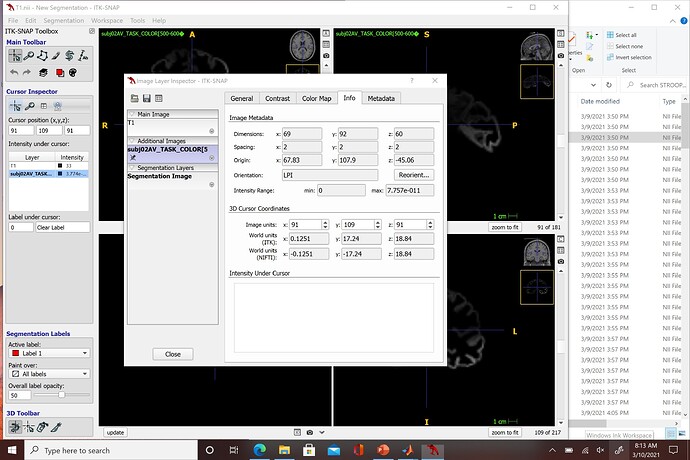
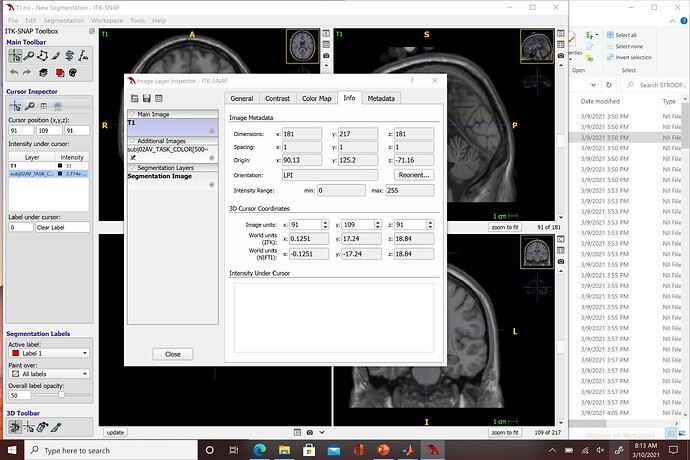
I am not sure if this is a similar issues that @tomasros was having in an earlier discussion but when I export my nifti files to SPM and attempt to run the stats I continue to get an error stating "The dimensions must be identical for this procedure" and "Failed: Factorial design specification". I am using the Colin27 T1 volume and ran it through ITK-SNAP to look at the voxel dimensions and it is 1:1:1. Unfortunately, all of the nifiti files I exported from Brainstorm are 2:2:2 which I don't believe SPM can read (is 1:1:1 in line with MNI?). I can change the voxel dimensions of each of my files in ITK-SNAP but it is quite tedious since I am working with 28 total subjects, each with 14 different nifti files of different time segment. Here are screenshots of the metadata from ITK-SNAP:
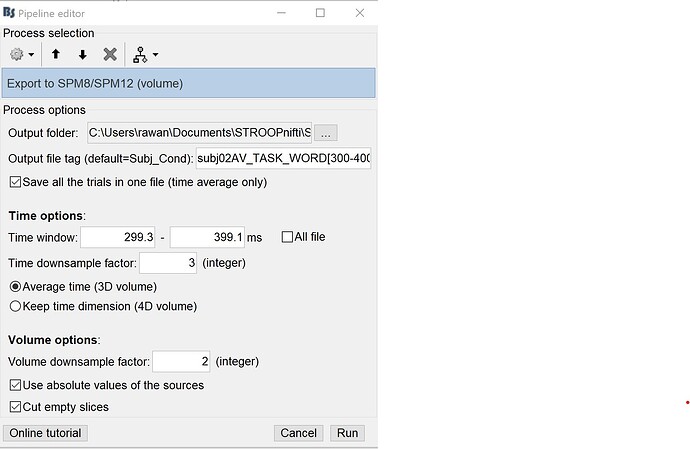
Is there a way to make this process more streamlined and set the voxel dimensions directly in Brainstorm before exporting my nifiti files? I've attached a screenshot of the properties I have set when exporting files. Based on your previous conversation it sounds like I need to uncheck the "cut empty slices" option and set spatial downsampling to 1 but I am unsure if this will fix my problem or not.
Thank you so much for you help,
Rawan
SPM should be able to handle files with 2x2x2mm voxels without any problem, but the problem is probably that you are mixing volumes with different dimensions.
The option "Cut empty slice" you does not save the slices that do not have any data (to produce smaller files), but this might mean that different slices from different files, and therefore that not all the volumes have the same size.
=> Disable this option to keep the original volume dimensions.
If this does not solve your problem, and that you actually have an issue with the voxel size: set the option "Volume downsample factor" to 1 instead of 2. Your files will be saved with a 1x1x1mm voxel size. But avoid it if you can, it creates unnecessary large files, as you can't expect to have a 1mm spatial accuracy from minimum norm source maps anyway.
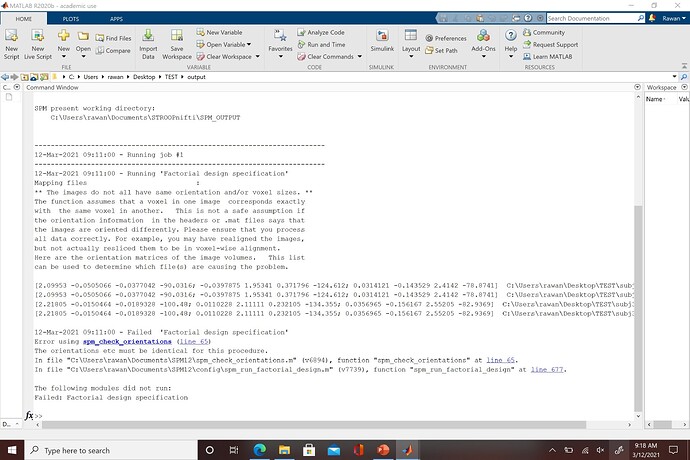
Thank you for the suggestions. I looked at the SPM manual again and you're right the 2x2x2 voxel spacing isn't the issue, it's the image dimensions. After I disabled the "cut empty slice" option my dimensions were identical across subjects when I checked it in ITK-SNAP. The issue that I am having now is that the origin dimensions are still different and SPM still cannot run the files. Could this discrepancy have occurred earlier in the pre-processing during configuration? Is there a way to fix it now or would it require going back to the beginning?
Different between what and what?
Since I fixed many bugs following the discussion with Tomas (Export to SPM: erroneous coordinates of origin in Nifti output file - #15 by Francois), make sure you update Brainstorm before trying anything new.
It appears that the image matrices for each of my subjects is different. Here is an example from the matlab error message with the first two matrices from one subject and the bottom two from another:
Is this because each subject had an individually warped MRI? If I didn't want to warp the MRI and instead just use the template (Colin27) across all subjects can I go back and redo my MNEs and spatial smoothing but with the template anatomy and then re-project the sources on the cortex before exporting them as niftis?
(Also, I updated Brainstorm per your recommendation)
Is this because each subject had an individually warped MRI?
You can't compare different source spaces voxel by voxel.
If you warped the template anatomy, then project the sources back to the template before doing any group analysis. Then all the exported source maps would be based on the same anatomy, and you wouldn't have any problem.
If I didn't want to warp the MRI and instead just use the template (Colin27) across all subjects can I go back and redo my MNEs and spatial smoothing but with the template anatomy and then re-project the sources on the cortex before exporting them as niftis?
If you want to use the template only: you need to start over from the very beginning and re-import all the recordings.
Use ICBM152 instead of Colin27.
If you have access to the digitized head points for each subject, the warped option is maybe better (if you aligned correctly the electrodes on the head...)