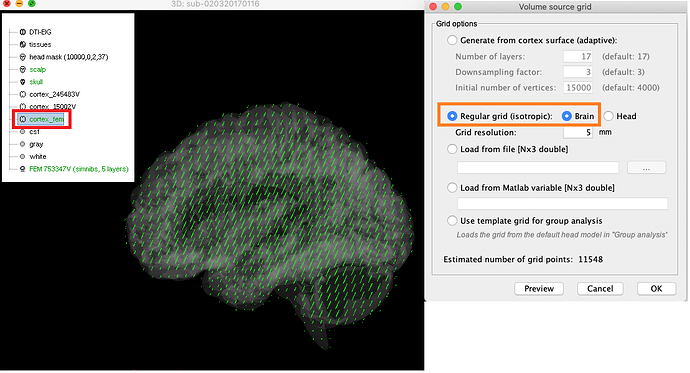
As I had issues with FEM headmodel and discussed with you under the thread, I am still attempting with different options, the issue I had before occurred when computing the headmodel with MRI volume -> head grid, the reason of doing it is that I'd like to extract time series from the deep structure after the inverse solution. Now I found the error is not shown if I change my option to MRI volume -> brain grid, however, the default setting from the anatomy is cortex 15000V which has only cortical regions. If I change it to cortex (FEM) that included deep structure(brainstem and cerebellum, etc) the preview looks good to me as shown in the attached figure.
My questions:
Is switching from cortex 15000V to cortex(FEM) an acceptable way of making headmodel?
If I selected with only the brain grid, does that mean the headmodel computation only uses the mesh of the inner layer without considering the conductivity of the rest four layers?
Would it be a good idea to force the source space to gray layer in this case(compute with brain grid) or it is no longer needed?
As introduced in the tutorial, it is recommended to remesh from simNibs FEM output, since there is no error reported, is it still a compulsory step to do before making headmodel?
Thank you very much for your time and looing forward your reply.
 . For it to work, you could try recompiling the code, depending on how comfortable you are with linux c/c++ development, it might be very easy or quite time consuming. There was a flag that needed to be activated in duneuro's settings script file. Duneuro has specific compiling tool that takes care of the packages. As I said, depending of how comfortable you are, it might be worth just to try to parallelize at the subject/paradigm/stimulus level through different calls to the serial application we distributed. If you want to dive deeper let me know. Take care,
. For it to work, you could try recompiling the code, depending on how comfortable you are with linux c/c++ development, it might be very easy or quite time consuming. There was a flag that needed to be activated in duneuro's settings script file. Duneuro has specific compiling tool that takes care of the packages. As I said, depending of how comfortable you are, it might be worth just to try to parallelize at the subject/paradigm/stimulus level through different calls to the serial application we distributed. If you want to dive deeper let me know. Take care,