Hello,
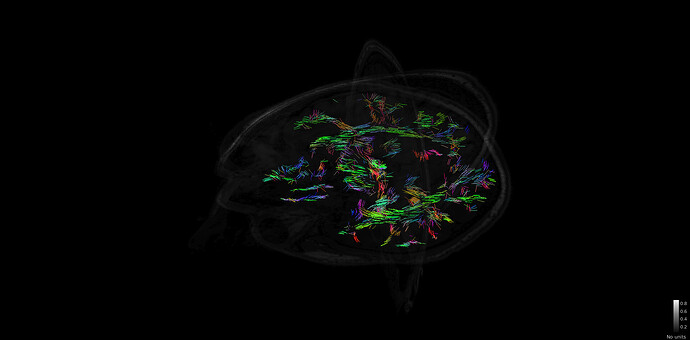
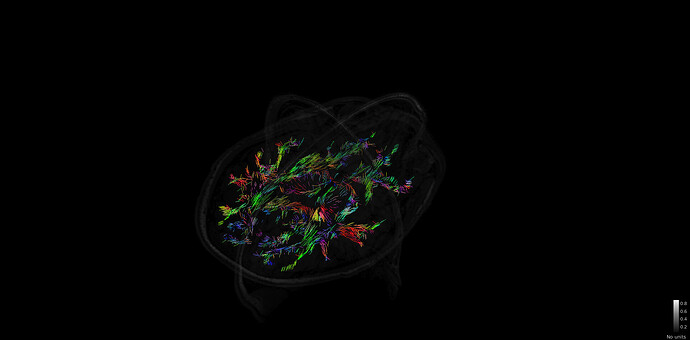
I just tried to reproduce the tutorial https://neuroimage.usc.edu/brainstorm/Tutorials/FemTensors on some of my data. But the FEM tensor estimation is very strange and it seems that all the fiber are only in one plane :
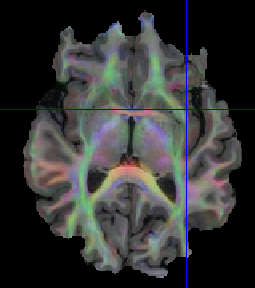
edit; it seems that when moving the plane, and forcing the redraw of the tensor, i indeed have fiber estimation for the entire brain, but i am still not sure if the fibers are ok.
How could I check it? I uploaded the data here and with the output from BrainSuite in the derivatives folder: https://drive.google.com/file/d/1ZUIqakHGMwqOBEEvllrUfqOCYWJxR0YP/view?usp=sharing
Also, how could I generate the fiber track to visualize them like in https://neuroimage.usc.edu/brainstorm/Tutorials/FiberConnectivity ?
Thanks a lot,
Hey @edelaire
You can open these files with Brainsuite as well; you can check here:
Currently, the fiber visualization is compatible with the TRK file format. We don't have the plugin to get the TRK file format within Brainstorm yet. So you need to process the DTI with third-party software that generates the TRK file, and then import it to Brainstorm as explained in the tutorial.
Thanks for your reply. I checked in BrainSuite and it looks fine:
I am a bit worried about some frontal region
is it expected to have this shape for the brain? is it an issue because of the B0 inhomogeneities or is it an issue with the brain mask?
When computing the tensor, I have this warning:
BST> Warning: 93 element(s) have max/min eigenvalue ratios > 10.
is it important?
Also, from the Brainsuite tutorial, i don't have the .odt file. Here are the files brainstorm generate: Should I run bdp outside of Brainstorm?
bse_cortex_file.nii.gz
great,
it's probably the quality of the MRI and then the segmentation.@Anand_Joshi any recommendations?
edelaire:
is it important?
No, that's just some information
that's actually a good value and close to the common value used for the WM conductivities.
I believe that the Brainstorm implementation does not call the function to generate the odt files.