Hello,
I am new to using brainstorm for fNIRS data processing. I want to inquire about how to import .hdr fNIRS file into brainstorm. I will appreciate a guide on how this can be done.
I look forward to hearing from you.
Thank you
Hello,
I am new to using brainstorm for fNIRS data processing. I want to inquire about how to import .hdr fNIRS file into brainstorm. I will appreciate a guide on how this can be done.
I look forward to hearing from you.
Thank you
Hello,
Which device did you used to get a .hdr file? For the moment, nirstorm only supports .nirs and .snirf. If you can share an example file with us; we can check if it is easy to support this new format.
Thanks for responding.
NIRScout was used to collect the data. I have provided a sample of the file extracted to this message
NIRS-2016-05-26_007.hdr (4.0 KB)
NIRS-2016-05-26_007_config.txt (376 Bytes)
Hello,
I just had a look at some data I acquired years ago in another lab, I have extra files that I think are needed: .wl1, and .wl2; .tpl .set .inf and .evt.
Do you have them by any chance ?
Also, if you can, I would suggest you to export the data as .snirf instead of native format
Yes, I have them. They are quite large (that was why I didn't upload it earlier). However, kindly find the link to the file (http://doc.ml.tu-berlin.de/simultaneous_EEG_NIRS/NIRS/VP001.zip)
How do I export the data as .snirf? The data is an open dataset so I am quite limited as regards how I can export the dataset.
Thank you for your help and I look forward to hearing from you.
oh i thought it was data you acquired. NIrx software allows you to do it. I found this converter online: NITRC: NIRx2nirs: A NIRx to .nirs data converter: Tool/Resource Info but it seems it requires a .sd file.
I worked with this dataset in the past; and tried to ask the information to the authors but they never replied to my emails...
If you want to analyse the data, my best suggestion is to download the .mat files and use the following script to import into brainstorm :
shin_import.m (8.7 KB)
You will need to replace lines at the begening to indicate the path to your data nirs_data_folder, eeg_data_folder, the number of subject you want to import n_subject and the name of the protocol in Brainstorm (protocol_name).
If I remember well what I did, it should import both EEG and NIRS (already as concentration).
In the mean time, I will process with the importation of the NIRx file in case you get access to this .SD file ![]()
Thank you so much!!! It worked perfectly well.
I just have one more comment on that.
I seem to be getting this error when I try to display the time series data. Do you have an idea of what I could be doing wrongly?
Hello,
I was able to reproduce the issue. it was an issue on how the unit were named. You can replace the line 123 of the import script from to sDataOut.DisplayUnits =data.oxy.yUnit; to sDataOut.DisplayUnits = strrep(data.oxy.yUnit,'/L','.l-1'); and then reimport the data. You will need to delete the Brainstorm protocol first, I think.
You can then visualize the signal:
One thing you can see however is that we don't have the position of the sensors in this dataset but only the mid-point between each source and detector. This information would be the .SD file mentioned earlier that the author should provide. I guess that with some guessing it would be possible to 'guestimate' some positions but that would be very approximative.
Note: If you right-click on the NIRS channel definition and do 'edit channel file', you can see for each channel, you can get the corresponding EEG name
Thank you edelaire,
Unfortunately, it is still not giving the necessary result. I have attached the script I used. Please do let me know if I am during something wrong. I have visualize the sensors and the NIRS channel. However, I am not able to visualize the time series. It seems to be giving me this error.
I look forward to hearing from you.
import_modified.m (8.9 KB)
I think this is an issue with Brainstorm (cc @Francois)
Update Brainstorm again and restart Matlab.
If you update Brainstorm from the Brainstorm interface, it works only if it is installed in a folder named brainstorm3. Otherwise, delete the brainstorm program folder and download a new one from the website.
Thank you so much!!!
It works now.
One more thing.... I keep getting errors when I perform operations like filtering. What can I do to rectify this?
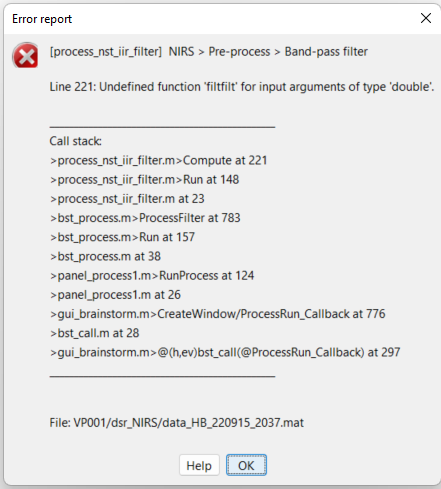
Indeed, we were relying on the matlab signal processing toolbox for filtering. I opened a pull-request (add oc_filtfilt by Edouard2laire · Pull Request #578 · brainstorm-tools/brainstorm3 · GitHub) to fix it.
This process is using IIR filter. Otherwise, you can use Brainstorm filter process that is using FIR filter. Just make sure to specify the transition band and check that the filter response correspond to the filter you want.
More info here: https://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#Filter_specifications:_Low-pass.2C_high-pass.2C_band-pass
Edouard
It works!
Thank you for your help
The PR is now merged in Brainstorm: add oc_filtfilt by Edouard2laire · Pull Request #578 · brainstorm-tools/brainstorm3 · GitHub
Update Brainstorm to get oc_filtfilt in the Brainstorm distribution.
Also update NIRSTORM ![]()
Thnk you for the update. I have updated it.
Just one more thing, when I try to use NIRS>Pre-process-Bandpass filter, it gives the error I've attached in my matlab window but still performs the operation. Should I be bothered about that?
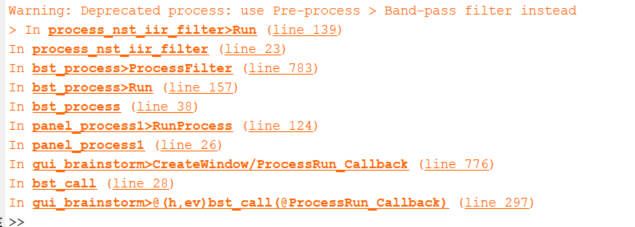
This error does not show up when I use Pre-process-bandpass filter directly as suggested