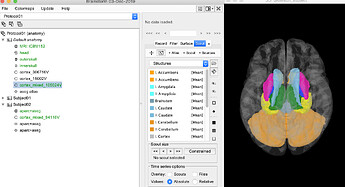
The default Brainstorm labelled aseg atlas (tess_aseg.mat) is a pleasure to use, however I need also the bilateral subcortical region "ventral DC" (region #28 in the screenshot under section 2 here)... Is it at all possible to have a version with it included? (In other words a tess_aparc+aseg.mat?)
Otherwise, is there some simple way to add it?
Grateful for any help!
You can load the APARC+ASEG.mgz volume as a volume atlas (Scout tab), or a set of surfaces (right-click on subject > Import surface).
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Volume_atlases
Hi @Francois, thank you for your directions! Unfortunately I still haven't been able to generate the figure I wanted... (sorry if I'm being obtuse!)
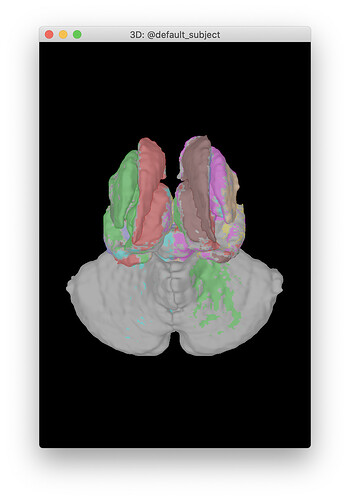
What I do want is for the end figure to look like is this, created by following TutorialDba (using tess_aseg.mat) but including the "L &R ventral DC"
Here is what I did (please tell me what I did wrong!) to import the aparc+aseg.mgz downloaded from Freesurfer:
- double clicked on "aseg atlas" under "Default anatomy". (Because otherwise the options within the "Scout" tab are not clickable...)
- Under "Scout" tab, chose "Load atlas" > aparc+aseg.mgz
(which file format amongst "Volume mask or atlas [...]" should I choose? The one that kind of works that I've tried is [no overlap, MNI space].) - It asks if I want to apply a standard transformation, so I say "Yes" and I get this:
but it's not like the wanted well-defined structures as when following TutorialDba....
Please help! ![]()
Copy the mgz file somewhere else, rename it to remove the "aseg" word, so it is not interpreted as an ASEG atlas by Brainstorm.
Then Import surface > File format = Volume mask or atlas (subject space).
Downsample the resulting atlas before displaying it (it contains > 1 million vertices)
In the Scout tab, the atlas Structures contains the meshes.
It might be more meaningful to compute volume source models and import your atlas as ROIs
Thank you so much @Francois it worked!! 
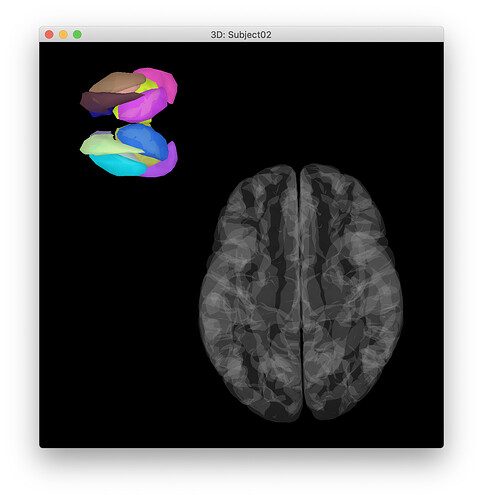
Hi @Francois , one further question: if I use tess_cortex_pial_low.mat as used in the TutorialDba, the subcortex doesn't go inside the "cortex" and I get an image as below -- how can I fix this?
Thanks again, your help is much appreciated! ![]()
This might get complicated to figure out how all the changes of coordinates systems are working...
I think the easiest would be to load everything from the same FreeSurfer output. Do no try to use the default anatomy (MNI ICBM152 template) at all: load the FreeSurfer folder for your subject (menu Import anatomy folder) and then add the additional atlas you want (Import surfaces).
Thank you so much for your help @Francois !!!