I'm using raw MEG data which I import to the database. I run the Hilbert Transform process on the data and can export this file easily to Matlab, which I've already successfully done. However, these files are unnecessarily big and of course cover the entire brain. I made a scout for the specific ROI and I want to run the Hilbert Transform specifically on the region this scout covers to avoid huge file sizes, but I don't see any clear options on the Hilbert Transform process window involving using scouts.
Hi Rachel,
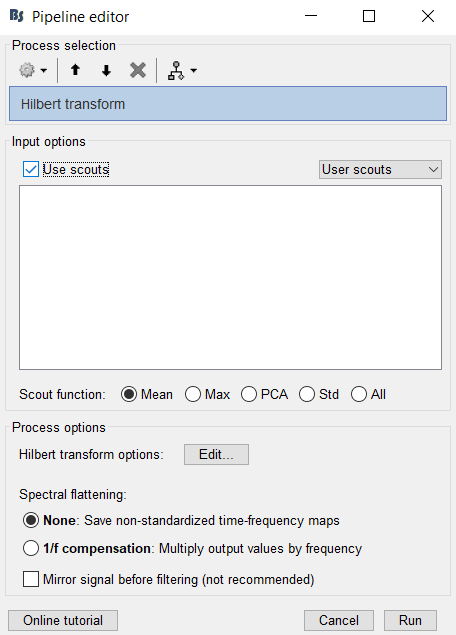
The Hilbert Transform process does have an option to choose which scout(s) to compute it on. Make sure that you are processing sources and not recordings in order to see this option. I am assuming you already have computed a head and source model.
Martin
Thank you! I was accidentally processing recordings which explains why the option wasn't there.
Now that I'm trying to run it on sources, I see the scout option but do not see the scouts I created in the User Scouts. I averaged the power maps from each trial for each Subject and was creating the scouts in User Scouts based on the average brain's alpha band map. I assume I need to project the scout to the individual brains to run the Hilbert Transform on them. I tried "project to: Default Anatomy" and on individual Subjects, but my scouts were still not listed as an option.
Thanks again for your quick reply!
From the Scout tab, you should be able to project your scouts defined on the default anatomy onto the each subject, using the menu Scout > Project to > Subjectxx > cortex
Make sure you select the correct surface...
If you still can't find a way to make it work, please post screen captures illustrating all the steps your are following
That was what I was trying to do, I must be missing some steps.
First, I created 3 scouts (10, 11, 12) in User Scouts on the averaged power maps for all the blocks from one Subject ( power maps obtained by following the processes in this tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/RestingOmega#Power_maps )
screen capture: https://drive.google.com/file/d/1HyrEpSoq4iDRtYmPUaxo0cLtHeMimxDJ/view?usp=sharing
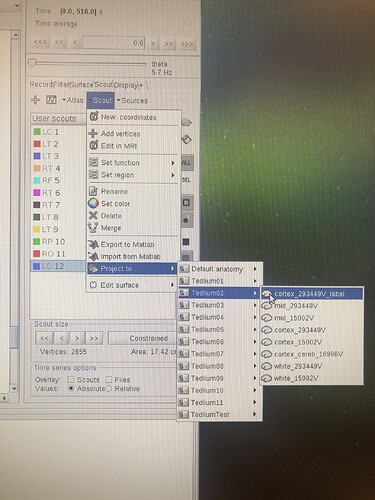
I then used the menu Scout > Project to > Tedlium02 > cortex(...)
Where "tedlium" signifies "subject". I tried all 4 cortex options pictured since I wasn't sure which might work.
After it was successfully projected, I reloaded the "Tedlium02" file, then tried to process the sources from that Subject with the Hilbert Transform which still did not have the new scouts (10, 11 or 12) as an option. (the usual options "right", "left" etc. are still there, can be seen on right side of screen capture)
Thanks for your help!
I don't know what is wrong with your procedure, what you describe looks OK.
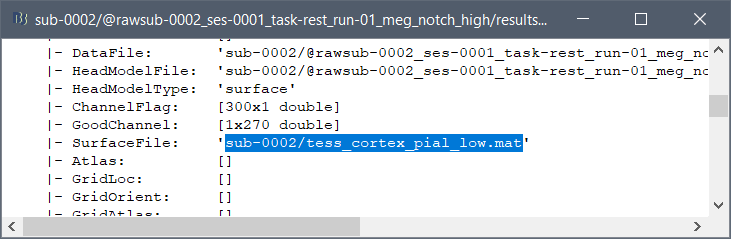
First, make sure you track correctly from which and to which surface files your are projecting your scouts. Typically, it would be the low-resolution cortex (cortex_15000V)
To check which file was used to estimate a source file, right-click on the file > File > View file contents, and look for variable "SurfaceFile":
Then hover your mouse over the various surface files in the subject anatomy to see which surface is the one that has this file name.
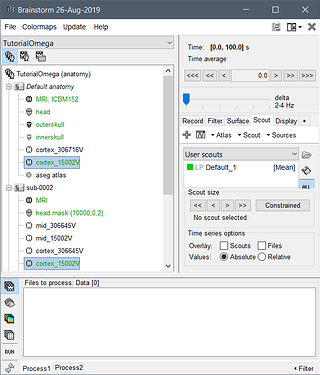
After projecting your scouts to Tedlium02/cortex_15000V (for instance):
- make sure there are no errors displayed in the Matlab command window (one possibility is that the procedure crashes)
- then double-click on the file Tedlium02/cortex_15000V and see what is available in "User scouts" (rather than trying directly the computation of something else)
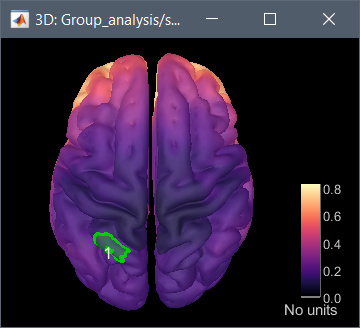
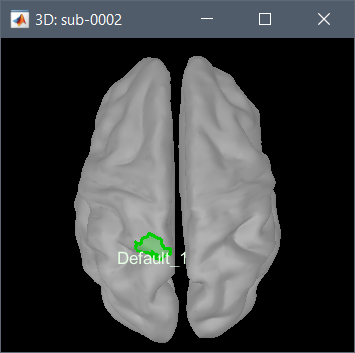
I tested it on the OMEGA tutorial dataset you pointed at, and it works as expected:
That worked perfectly, again thank you very much for your help and quick responses!