Hello
Thank you for the training you provided for brainstorm. Regarding the line related to the head model that is in the FildTrip : (cfg.headmodel = headmodel_fem_eeg_tr; % the head model), I would like to get help to extract this head model matrix from where in the brain storm. Thank you for your response.
In the GUI, you can right-click on the head model (![]() ), then File > Export to Matlab. This is will create a variable with the head model data as described in the link below
), then File > Export to Matlab. This is will create a variable with the head model data as described in the link below
Otherwise, you can import that data from head model file with the function: in_bst_headmodel
HeadModel on the hard drive:
https://neuroimage.usc.edu/brainstorm/Tutorials/HeadModel#On_the_hard_drive
Thank you for your response. I wanted to use a method other than the ones available in BrainStorm, so I was forced to extract the file from BrainStorm. For using FieldTrip according to its instructions, I need the headmodel and lead field. Based on the extraction method described in the tutorials you mentioned earlier, I am extracting the file and using gain as the lead field.
However, I don't know which parameter to use for the headmodel. According to the instructions in FieldTrip:(cfg.headmodel = headmodel_bem; % the head model
cfg.grid = leadfield_bem; % the precomputed leadfield), could you please explain the difference between the head model and lead field, and the values that are used for each? Thank you for your knowledge and the time you're dedicating to me.
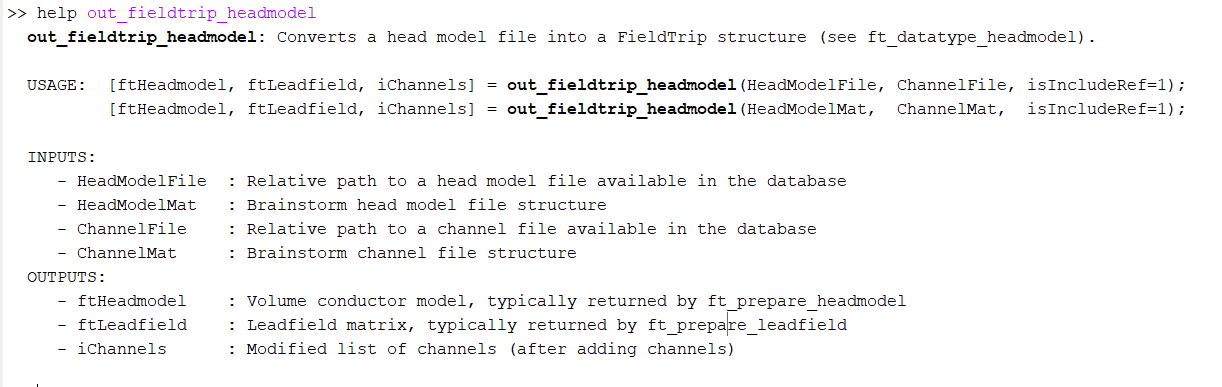
In Brainstorm, there is the function out_fieldtrip_headmodel() that will create the FieldTrip headmodel and leadfield, from a Brainstorm head model file (note that in Brainstorm the leadfield information is inside the head model file).
brainstorm3/toolbox/io/out_fieldtrip_headmodel.m at master · brainstorm-tools/brainstorm3 · GitHub
If you want to go the opposite way, form FieldTrip headmodel+leadfield to Brainstorm headmodel, check the code in the process_ft_prepare_leadfield. With that process, it is possible to compute a headmodel-leadfield pair with FieldTrip and save it as a head model file in Brainstorm.
The are very much related but they are not the same:
-
Head model: refers to the representation of the head tissues, such as white and grey matter, cerebrospinal fluid, skull bone, and skin. This is also know ad volume conduction model and specifies how source currents in the brain are propagated through the tissues. The head model is independent of the sensor positions, only depends on the head anatomy and geometry.
-
Lead field: is the one also know and forward model, is a matrix that relates the activity of the sources in the brain to the sensor data. For it's computation, the head model and the sensor locations are needed.
Thank you very much for the helpful guidance. I have a question - I am using a GUI and I would like to know how I can use this function. ( [brainstorm3/toolbox/io/out_fieldtrip_headmodel.m at master · brainstorm-tools/brainstorm3 · GitHub ])to extract the headmodel and leadfield from brainstorm?
hey @ttaleei
you can quickly check the help of this function, and you have all the required inputs explained.
Thank you very much
Hello.
I am new to the Brainstorm software. I apologize for my simple question. I created new data through simulation with SimMEEG. Could you please guide me on the difference between the HeadModelFile and the HeaderModelMat as well as the difference between the ChannelFile and the ChannelMat, and from which part they are extracted in the Brainstorm? Thank you for your knowledge and the time you're dedicating to me.
The ones ending in File are the path to the files in the database.
The ones ending in Mat are the content of such a files.
If you are a new Brainstorm user, we recommend you start by following the introduction tutorials first (section "Get started" on the tutorial page), using the example dataset that is provided. This will get you familiar with Brainstorm data.
For scripting, check this tutorial. But only after the introduction tutorials, as it relies a understanding of concepts in Brainstorm
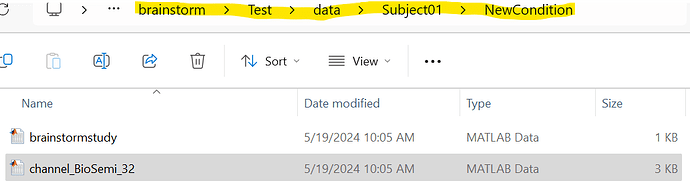
Thank you very much for the time you have provided me. Given the explanation you provided, the channelFile path is the location where the mat file is placed, so for the channelFile I should place the highlighted parts inside the image, and for the mat file it is the file that is in the path (channel_BioSemi_32),
and for the HeadModelFile and HeadModelMat it is the same.
Is that correct?
@ttaleei, you can run the function only with the file names, I do not why you want to load them.
Check the first line of the usage provided above. Please read carefully the information provided
The path to is the entire path. This is to say absolute path for folder, filename and extension
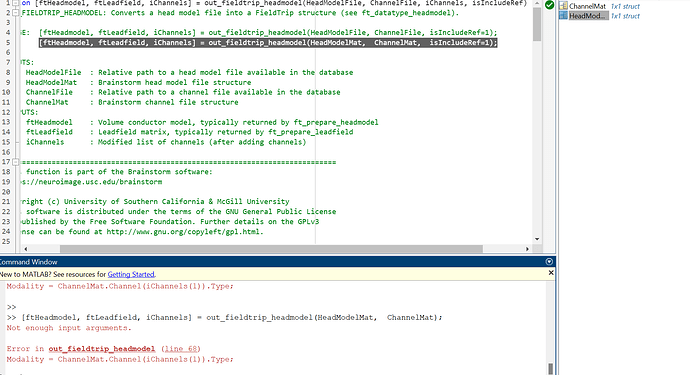
Thank you for the quick response. I'm confused - in order to run the function, I need to provide the path to the channelFile and the headModelFile, but when I do that, I'm encountering an error (Unable to use a value of type string as an index.
Error in out_fieldtrip_headmodel (line 68)
Modality = ChannelMat.Channel(iChannels(1)).Type;) because I don't have the iChannels. Could you please guide me on what to do, as I'm very confused.
Can you share what arguments you are using for that function?
It's clear from the help that you can use either the Matfile loaded to Matlab workspace or the absolute path where these files are saved.
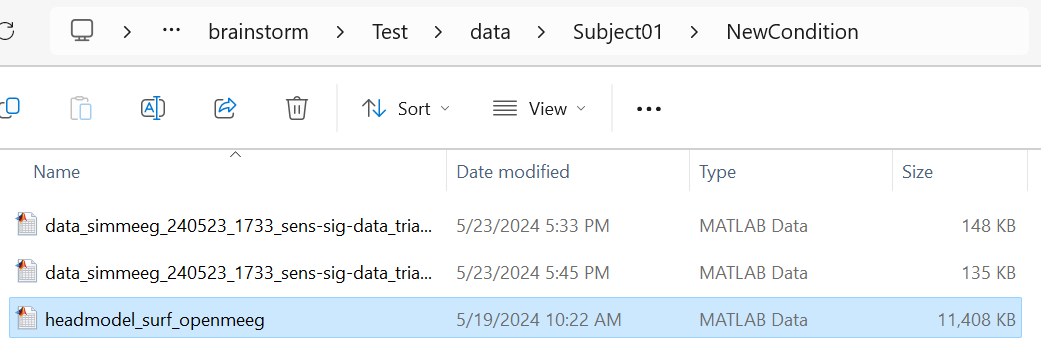
I have saved and loaded the channel_BioSemi_32.mat and headmodel_surf_openmeeg.mat files into the MATLAB workspace, but I'm encountering an issue with the iChannels parameter in the [ftHeadmodel, ftLeadfield, iChannels] = out_fieldtrip_headmodel(HeadModelFile, ChannelFile, iChannels, isIncludeRef) function. The function documentation does not explain the expected format or purpose of this parameter, and when I execute this function:[ftHeadmodel, ftLeadfield, iChannels] = out_fieldtrip_headmodel(HeadModelFile, ChannelFile, isIncludeRef=1);, I'm receiving the error that mentioned above(Unable to use a value of type string as an index.
Error in out_fieldtrip_headmodel (line 68)
Modality = ChannelMat.Channel(iChannels(1)).Type;) .
Thank you.
The function needs only two parameters, and you are using more than that
I attempted the procedure shown in the picture below, but I encountered an error.
[ftHeadmodel, ftLeadfield, iChannels] = out_fieldtrip_headmodel(HeadModelMat, ChannelMat);
Could you assist me further?
Thank you.
can you try by adding 1 as third argument,
[ftHeadmodel, ftLeadfield, iChannels] = out_fieldtrip_headmodel(HeadModelMat, ChannelMat, 1);
I am very grateful for answering all my questions and helping to resolve my issue.
As per the code you provided above, I used 1 : 32 instead of just 1 for 32 channel labels.
[ftHeadmodel, ftLeadfield, iChannels] = out_fieldtrip_headmodel(HeadModelMat, ChannelMat, 1:32);
Thank you.
I want to move the channels out of Brainstorm and over to FieldTrip, and have a file with the following specifications. Could you please provide some guidance?
Indeed, the help string for out_fieldtrip_headmodel is outdated, we will fix ASAP.
That missing argument is in fact the index for the channels that you want to use for the leadfield computation. In your case as there are only 32 EEG channels, using 1:32 is ok.
![]() We encourage you to explore the Brainstorm documentation, GitHub repo and Forum for similar questions, as the answer is often few clicks away. For example, the answer for this question can be found:
We encourage you to explore the Brainstorm documentation, GitHub repo and Forum for similar questions, as the answer is often few clicks away. For example, the answer for this question can be found:
-
In the links that we shared earlier state how to Convert Brainstorm structures to FieldTrip structures:
https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting#Reference:_File_manipulation -
By searching in the Forum, this will lead you to this thread with the required information:
Convert BS to filedtrip