No, indeed.
You need to find a way to project your atlases on the individual brains first, and then import them into Brainstorm. I don't know how to do that, but there are probably ways with FreeSurfer.
I've just been told by Chris (Markiewicz) freesurfer's mri_surf2surf function might be doing it but haven't had a chance to have a look. Once I get more info and practices I will report here but for now several links about the function itself is here
https://surfer.nmr.mgh.harvard.edu/fswiki/mri_surf2surf
And I guess Jeff was asking the same question in freesurfer forum so here is the answer he got regarding mri_surf2surf function
https://www.mail-archive.com/freesurfer@nmr.mgh.harvard.edu/msg34517.html
I will let you guys know if I can get any improvements but it might take time.
Thank you
Isil Bilgin
Thanks for reporting all your findings here, this will definitely help other people.
Hopefully you'll manage to have CAT12 working, it would be an easier solution.
Hi Francois,
I feel I am missing a step.
I do the following:
new sub: test01
import MRI
import surf: /surf/lh.pial
display lh.pial
Scout:Load Atlas:Load Atlas /label\lh_Yeo.annot
close and clear
This works great.
Now I have the Yeo atlas on the lh.pial surface.
And it looks good.
I then need to downsample the lh.pial to 7500 vertices.
Right click lh.pial => less vertices
Once I have downsampled lh.pial the atlas no longer appears to fit.
Its there on the surface but it looks like a terrible fit.
Am I missing something?
Do I need to also downsample the atlas?
How do I do this in Brainstorm?
Thank you.
-Tom.
The support for importing automatically the Yeo cortical parcellation was added to the FreeSurfer import function. If you have files named 'lh.yeo2011_7networks_n1000' or 'rh.yeo2011_7networks_n1000', they should be added automatically as an atlas labelled "Yeo 7 Networks".
Same for the 17 Networks.
https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/io/import_label.m#L696-L699
Instructions to compute these files can be found here:
https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#Cortical_parcellations
However, I recommend you use the Schaefer atlas from CAT12 instead (the updated version :
https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12#Cortical_parcellations
Thanks Francois.
Briefly, the problem we are having is this.
We are using Desikan with 68 regions but we only have 62 electrodes.
This means after source-localization some of the 68 are not independent time series.
In order to perform leakage-correction (orthogonalization, all at once, not pairwise), we need to have 68 independent time series.
Hence, we presumed we needed to use an atlas with fewer regions.
The Schaefer seems to have many regions.
The Yeo 17 has 17 networks. This means (i think) that network 1, for example, will have multiple regions spread across the cortex but only provide a single time series for that network.
This is not quite optimal for our purposes.
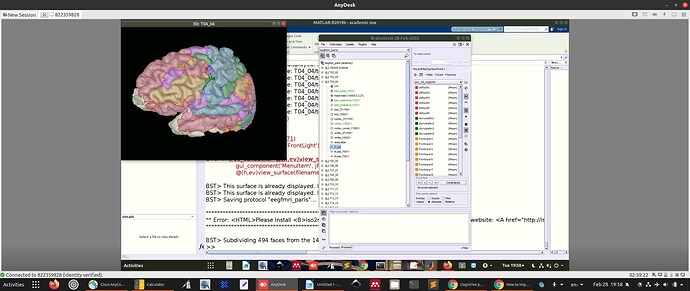
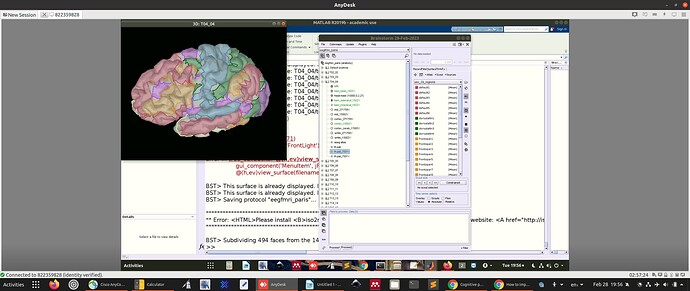
For the above reasons, I have imported a version of the Yeo with 29 regions per hemisphere (composed of the 7 networks).
You can see the 29 individual regions in the picture above.
I trust this comports with your own understanding?
I was just a bit concerned that the downsampled atlas has somewhat of a patchy appearance.
But maybe this is normal?
Apologies for the long explanation.
Thanks Francoise.
We were basing our assumptions on this:
Paraphrasing:
The issue you are now facing relates to earlier warnings from @GilesColclough; the atlas has too many regions given the linear rank of the underlying data. The reason that the data has "low" rank in the first place (keeping in mind that 100+ dimensions is not actually that low!) is due to the ill-posedness of source-reconstruction in MEG. Basically: we are trying to estimate 3k+ signals (one for each voxel) from the correlated measurements of only ~250 sensors, and as the error message says, this gives us only 114 linearly independent signals to work with --- but the AAL parcellation assumes the existence of 115 independent regions.
There are essentially two ways forwards:
- If the AAL parcellation is not a must for your analysis, then consider using a parcellation with fewer regions (e.g. Desikan-Killiany);
- Otherwise, I would first remove all subcortical regions from the parcellation (the SNR in these regions is extremely low compared to cortical regions), and perhaps merge small regions together, or even with larger neighbours. For example, find parcels with very small label counts after the resampling, and either merge neighbours with small counts together, or else merge small regions with their smallest neighbour.
To clarify, we get 68 regional timecourses (Desikan atlas) and perform orthogonalizatoin (Colclough et al. 2015). And the algorithm fails with the message:
The ROI time-course matrix is not full rank.
This prevents you from using an all-to-all orthogonalisation method.
Your data have rank 60, and you are looking at 68 ROIs.
You could try reducing the number of ROIs, or using an alternative orthogonalisation method.
We search the 68 time series for those that are linearly independent.
We keep those and remove the rest.
We now have ~62 time series (depending on the subject).
We run the orthogonalization alogithm again.
This time it is successful.
So i'm not sure I understand.
Not all the timeseries seem to be contaminated with each other because we can remove them and have the algorithm work.
We therefore assume that reducing the number of regions of the atlas will ameliorate this issue.
Please help me understand why I am wrong.
PS. Maybe you mean we should use a different solution to minimum norm?
First, I'd like to apologize for my previous inaccurate reply: the rank of the scouts times series matrix is indeed linked with the number of sensors.
If you have Neeg=62 electrodes, the rank of your recordings is at most 62, or 61 if the data is in average reference and the recording reference is included. Additionally, the rank decreases for each SSP or ICA component that you remove during the signal cleaning.
Each source signal is a linear mix of the Neeg EEG signals. If you consider a set of more than 62 sources, the matrix [Nsources x Ntime] may have a rank up to Neeg. The scouts time series are averages of source time series, and would therefore share similar properties. When computing the signals associated with more than 62 scouts, the resulting matrix [Nscout x Ntime] may have a rank up to Neeg.
Limiting the number of scouts to 58 scouts would make it possible to obtain a "scouts matrix" [Nscouts x Ntime] that has a full rank. However, I'm not sure I understand this convoluted objective of computing scout signals and then orthogonalizing them:
- The orthogonalized signals would not have any correspondence with the underlying anatomical ROIs.
- Wouldn't you obtain similar signals as when processing the EEG signals directly? (minus the information that is lost during the inverse model computation - see regularization)
Working with volume sources would bring you closer to the pipeline you refer to, but then you would face a problem of having sources with unconstrained orientations (3 signals per ROI).
On the surface, the selection of the type of parcellation to be used is not trivial, and I don't know how to guide you with this. One recommendation I can give you is not to try using a volume parcellation (eg. AAL) for processing sources estimated on a surface.
As a software engineer, I am not competent to address these questions. I would recommend you seek advice from signal processing specialists.
@John_Mosher @Sylvain Can you please share your recommendations?
I was just a bit concerned that the downsampled atlas has somewhat of a patchy appearance.
Only vertices are classified as part of scouts, not faces. The faces between two ROIs are not attributed to one or the other and therefore not painted. Display the surface edges and zoom in to observe this.
Other threads discussing rank of source space signals:
François provides useful advice here.
In addition, if you truly want to work with as many regions as dimensions in your scalp signals, you can decide to merge some of the atlas regions together. This is a feature available in the Scout panel: select two or more scout regions from the list and select 'Merge' from the panel pulldown menu. Repeat until you reach the targeted number of regions.
Hi Francois et el.,
I am back on this thread again ![]() . I want to use the Yeo (Schaefer) atlases again, and read through this entire thread just now. It seems the quickest way to do this is to somehow use the CAT12 tool. However, when I followed the link, I get the BSt documentation that describes the process for computing scouts for individual brains. I just want the {.annot} files so I can create the generic atlas like I did 11 years ago. Where can I get those directly? Or can this CAT12 program accomplish that?
. I want to use the Yeo (Schaefer) atlases again, and read through this entire thread just now. It seems the quickest way to do this is to somehow use the CAT12 tool. However, when I followed the link, I get the BSt documentation that describes the process for computing scouts for individual brains. I just want the {.annot} files so I can create the generic atlas like I did 11 years ago. Where can I get those directly? Or can this CAT12 program accomplish that?
-Jeff
P.S. I found them here:
which has a direct link to just the files here:
I downloaded and unZipped the Parcellation.zip file and found the annots I think I want to use.
I tried to replicate what I had done in 2013:
create new protocol: Bu_FSAve
create new subject: Bu
under new protocol have
(+) Default anatomy
(+) Bu
r-click Default anatomy
l-click import surfaces
choose llh.Schaefer2018_1000Parcels_17Networks_order.annot
get message:
() MRI orientation was nonstandard and had to be reoriented
() apply the same transformation to the surfaces?
() default answer is NO
I tried it with both YES and NO
both times got this:
() file format could not be detected automatically
() please try again with a specific file format
Please advise.
-Jeff
If you run CAT12, you will have the Yeo / Schaefer atlases in Brainstorm.
To have the .annot file you could just export the Brainstorm atlas as such:
Select all the Scouts for the atlas, then click on the (![]() ) Save selected scouts to file ) and choose
) Save selected scouts to file ) and choose .annot
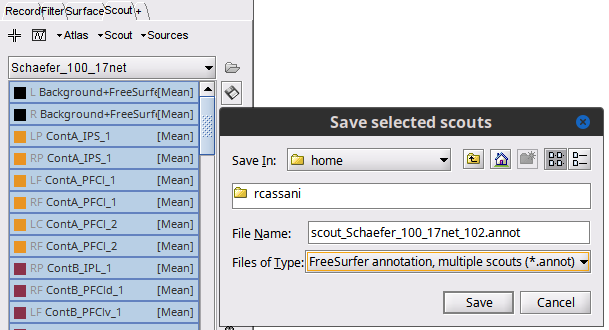
Raymundo,
I believe you, but really do not want to load up SPM and CAT12 just for one Atlas. Can you try to tell me what is wrong with the method I am trying?
-Jeff
.annot are not surfaces, they are collections of labels for a given surface. This is way they cannot be added as surfaces.
You can load in Brainstorm add the surfaces in the shared repository. Besides the annot files you need to surface files that they correspond to, e.g., lh.pial and rh.pial
- Create a Subject using the FsAverage anatomy (right-click on the subject Use template > FsAverage)
- Import left hemisphere surface (right-click on the subject Import surface, use All surface files(.) and select
lh.pialandrh.pial). Select YES to apply the same transformation. - Add Scouts for left hemisphere. Double-click the lh.plial surface, go to the Scout tab, Atlas > Load atlas..., select the lh.*.anot file. Close figure.
- Repeat step 3 for right hemisphere
- Merge hemispheres, select both surfaces, right-click Merge surfaces. This will create a new cortex_xxxV this is a surfaces with both hemispheres and the labels as Scouts.
Note that these surfaces and labels are only for surfaces in the repo (which are the FsAverage template). They are not for individual brains as in was mentioned in your question. While it is possible to project scouts from one surface to another, betters results are obtained if the labels are obtained during the MRI-segmentation process
Thanks for clarifying all this. I now see that the main problem is that I did not follow my own notes from July 2023 at the beginning of this thread. I somehow glossed over importing the pial surfaces and tried to load the annot files first.
I should be able to do this now with no problem.
A new problem to be solved for my purposes. After importing the Schaefer Atlas into the FSAve brain, I went to "Compute the head model". I got the warning that I do not have the skull and scalp surfaces. Well, yeah, they did not come with the pial surfaces, and I doubt they exist since this is an "average" pial surface and there might not be an "average" set of skull/scalp surfaces to match it. I will still try to look for one in the FS docs, but assuming they do not exist, can you suggest a possible strategy for mapping the FSAve surface to the ICBM surface along with the Schaefer labels? That seems like the best way to get what I need.
You could generated head, BEM and FEM surfaces using the MRI in the FsAverage template, check the MRI segmentation menu as suggested in here:
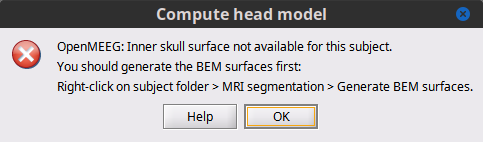
Ah, thank you. Now I get asked if I want to use BSt or FS mesh generation.
Check the BEM tutorial for more info about this options
https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem
P.S. Please create a new thread for new questions, as head model is not really related to importing brain atlases as scouts
OK, I will follow this up on a new thread.