Am I loading the mask correctly? If yes, what should be done to parcelize it according to broadman areas?
There is an issue with the registration between the MRI and the mask, you need to address that first.
https://neuroimage.usc.edu/brainstorm/News#MRI_coregistration
Regarding the Brodmann parcellation, it is cortical parcellation and the mask is volume information.
What is it that you are aiming to do?
I have the .nii files for 7 resting state networks. I want to parcelize these resting state masks into Broadmann areas and than create the corresponding binary maps. How can I do that?
Thank you for your response. I have 3-D mask of dimension 91x109x91 for different resting state network. I want to get current density value for all volume using sLORETA and perform piecewise multiplication between the binary 3D mask and the projected signals obtained from sLORETA. How do we decide the dimensions of the projected signals?
Thanks for the clarification. Once you have computed the source using a volume source space. It is possible to import the .nii masks as Scouts, and these scouts will indicate which indices to mask on the source reconstruction.
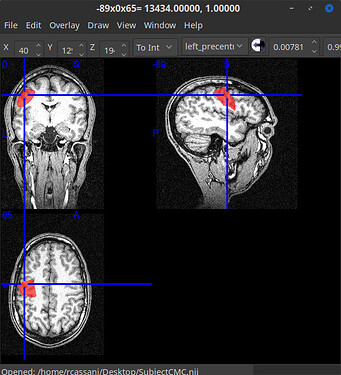
MRI and a mask (left motor cortex)
Scout created with mask.nii file, and its (236) indices, which indicate the source vertices in the Scout.
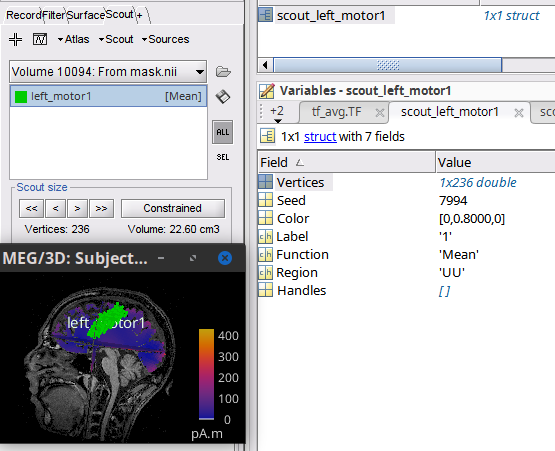
This is when the source space is defined, so the number of vertices.
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource
Hi , Thank you for your prompt response. I will surely try the suggested solution. But I have a few queries.
Q.1 Initially, I was trying to project sources to the default anatomy and then perform the pointwise multiplication in matlab with the .nii mask. Is this the right approach if we want to get current density value for all volume using sLORETA and perform piecewise multiplication between the binary 3D mask and the projected signals obtained from sLORETA.
Q.2 If The above approach is correct, I am still getting an error "Dimension exceeds" while performing project sources. How can I resolve that?
Q.3 In brainstorm, where can we get the current density value or source time series ?
You need to project the volume source estimation to the anatomy for which the mask.nii file is related. As mentioned in some post above, first verify that the mask and the anatomy are properly registered. Once this is done, you need to import the mask NIfTI file as a Scout.
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Volume_atlases
Please provide more details in the error that is shown, and what are the steps to get it.
This are in the sources file, that can be full results (![]() ) or a link (
) or a link (![]() ).
).
The time series can be obtained with the function:
sResults = in_bst_results(SourceFile, 1);
The 1 indicates that if the sources are a link, it needs to be resolved.
More info on the structure of sResults in here: https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#On_the_hard_drive