Dear colleagues,
I wish to run resting state analysis on my MEG data using brainstorm.
In this regard, I tried to processed the cited open MEG dataset (version 2) using the script published in this paper "Brainstorm Pipeline Analysis of Resting-State Data From the Open MEG Archive" available at https://www.frontiersin.org/articles/10.3389/fnins.2019.00284/full but I ran into problems.
bst_process('CallProcess', 'process_import_bids') processes the dataset for quite some time but returns back with an empty structure. So the rest of the analysis is not done.
How do I go about reproducing the resting state analysis results published in this paper?
Reference:
Niso, G., Tadel, F., Bock, E., Cousineau, M., Santos, A. and Baillet, S., 2019. Brainstorm Pipeline Analysis of Resting-State Data From the Open MEG Archive. Frontiers in neuroscience , 13 , p.284.
Many thanks,
Pradeep
Auditory Cognition Group,
Medical School, Newcastle University,
Newcastle Upon Tyne, NE2 4HH, U.K.
Website: https://www.auditorycognition.org/pradeep
I checked the processing script for this dataset, and it still works.
As recommended in the article, my advice would be that you use an updated version of Brainstorm, together of an updated version of the processing script.
- Delete all the existing version of Brainstorm
- Remove anything that is not related with Matlab itself from your Matlab path
- Restart Matlab
- Download Brainstorm from the website
- Use the script brainstorm3/script/tutorial_omega.m from this newly downloaded package:
tutorial_omega('/path/to/ds000247')
If you put a breakpoint at line 66, after the execution of process_import_bids, you should have a structure with the relative path to the all the links to the imported datasets. You can ignore all the warnings that appear in between.
K>> sFilesRaw.FileName
'sub-0002/@rawsub-0002_ses-0001_task-rest_run-01_meg/data_0raw_sub-0002_ses-0001_task-rest_run-01_meg.mat'
'sub-0003/@rawsub-0003_ses-0001_task-rest_run-01_meg/data_0raw_sub-0003_ses-0001_task-rest_run-01_meg.mat'
'sub-0004/@rawsub-0004_ses-0001_task-rest_run-01_meg/data_0raw_sub-0004_ses-0001_task-rest_run-01_meg.mat'
'sub-0006/@rawsub-0006_ses-0001_task-rest_run-01_meg/data_0raw_sub-0006_ses-0001_task-rest_run-01_meg.mat'
'sub-0007/@rawsub-0007_ses-0001_task-rest_run-01_meg/data_0raw_sub-0007_ses-0001_task-rest_run-01_meg.mat'
'sub-emptyroom/@rawsub-emptyroom_ses-18901014_task-noise_run-01_meg/data_0raw_sub-emptyroom_ses-18901014_task-noise_run-01_meg.mat'
'sub-emptyroom/@rawsub-emptyroom_ses-18901015_task-noise_run-01_meg/data_0raw_sub-emptyroom_ses-18901015_task-noise_run-01_meg.mat'
'sub-emptyroom/@rawsub-emptyroom_ses-18910228_task-noise_run-01_meg/data_0raw_sub-emptyroom_ses-18910228_task-noise_run-01_meg.mat'
'sub-emptyroom/@rawsub-emptyroom_ses-18910303_task-noise_run-01_meg/data_0raw_sub-emptyroom_ses-18910303_task-noise_run-01_meg.mat'
'sub-emptyroom/@rawsub-emptyroom_ses-18910512_task-noise_run-01_meg/data_0raw_sub-emptyroom_ses-18910512_task-noise_run-01_meg.mat'
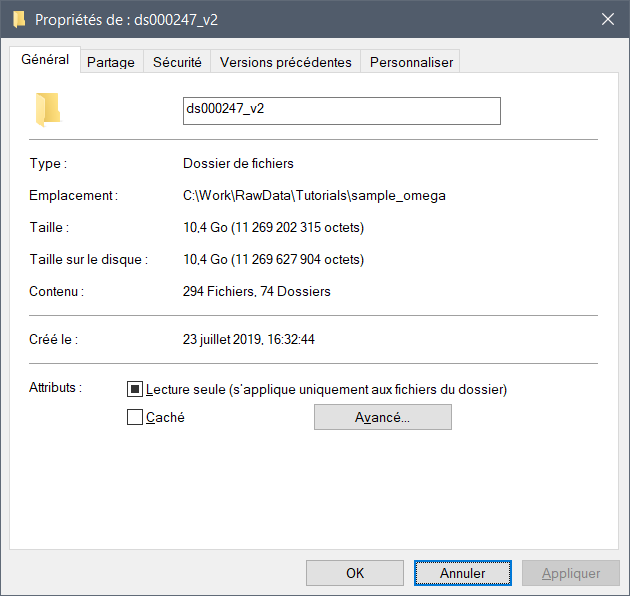
Otherwise, the dataset that you have downloaded might be incomplete. It should be over 10Go:
Thanks Francois.
After I provided the path of SPM installation in Brainstorm preferences, process_import_bids ran fine.
Now the script works fine until it hits "Process: Compute sources [2018]" that uses 'minimum norm' where it returns with an empty structure.
Do I need to install any specific toolbox in addition to SPM, Fieldtrip? How do I go about reproducing the resting state analysis results published in this paper?
Many thanks,
Pradeep
Auditory Cognition Group,
Medical School, Newcastle University,
Newcastle Upon Tyne, NE2 4HH, U.K.
Website: https://www.auditorycognition.org/pradeep
Do I need to install any specific toolbox in addition to SPM, Fieldtrip?
No, you don't have to install anything else.
How do I go about reproducing the resting state analysis results published in this paper?
By running the script tutorial_omega.m:
brainstorm3/toolbox/script/tutorial_omega.m at master · brainstorm-tools/brainstorm3 · GitHub
Or by doing it manually, as instructed in the online tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/RestingOmega
Maybe something changed in Brainstorm since it was published, or something is set up in a wrong way on your computer.
To get the message that caused the error: check the execution report after it crashes. Copy-paste the error messages here if you don't understand them.
(menu File > Report viewer, or bst_report('Open','Current'))
I found the issue. Since I had retained only 1 subject from your dataset but all five empty room recordings, the script was not associating any of the five noise covariance matrices with the subject causing the failure of source localisation module.
Now that I am able to reproduce the result on data set published in the paper, I am using my own resting state MEG data but I do not have empty room recordings, nor head shape points. I could follow steps listed for GUI to process the data. But I get the following error while trying to project the psd values onto the cortical surface.
One surface has an atlas "Structures", the other does not. You need to use surfaces coming from the same software to project sources.
How do I resolve this?
Best,
Pradeep
One surface has an atlas "Structures", the other does not. You need to use surfaces coming from the same software to project sources.
How do I resolve this?
You need to find out why one of your cortex surface has a "Structures" atlas and not the others.
This atlas is present in the anatomy template ICBM152 and in all the anatomy folders imported from FreeSurfer, CAT12 or BrainSuite.