Hello,
I recorded EEG using EGI amplifier, but recorded other physiological modalities using Biopac.
They are properly synced, but saved in separate file, different format.
Can I merge ecg data to eeg in certain step to remove ECG artifact?
You can link the two files separately, detect the artifacts in the files that contain the ECG, and then transfer them to the file with the EEG signals.
This is illustrated in this tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/EyetrackSynchro
@Francois Thanks so much!
It seems brainstorm cannot import .acq (Biopac Data Format) directly yet.
The list doesn't contain .edf for biopotentials, but shall I convert .acq to edf then select in 'review raw file'?
It looks like FieldTrip has a function that reads this file type: fieldtrip/load_acq.m at master · fieldtrip/fieldtrip · GitHub
If this works, I could add the direct support for these files in Brainstorm easily.
Could you please share one or a few example files?
- Zip the .acq files
- Upload the zip file somewhere (e.g. google drive)
- Post the download link here
In the meantime, yes, you could try converting these files to EDF. Keep in mind that it is never recommended to do too many data conversions, as there is always some info that disappears along the way.
@Francois
Fantastique!
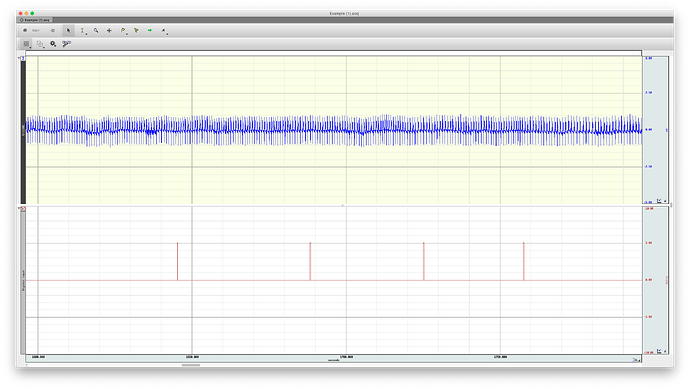
Below, a biopac .acq file recorded using Acqknowledge 5.0.2. one ECG channel and one event TTL channel.
I added the BIOPAC .acq format to Brainstorm:
https://github.com/brainstorm-tools/brainstorm3/commit/49493cf77f4d48e306945ceae16f6582be386555
You example file looks empty. Is this expected?
@Francois
No it's not an empty file.
It should contain one ECG channel and two TTL signal channels (might be looks empty).
The function load_acq reads two signals from this file.
>> acq = load_acq('C:\Work\RawData\Test\biopac\Example.acq')
Loading C:\Work\RawData\Test\biopac\Example.acq .. Done!
acq =
struct with fields:
hdr: [1×1 struct]
data: [2162690×2 double]
Signal 1 = 2.5 everywhere
Signal 2 = 0, except for a few samples at the beginning.
If this is not correct, then I'm not sure how to read this file in Matlab...
Do you have access to other Matlab functions that can decode this file?
@Francois
As far as I know, the only working solution for Biopac .acq is using acq2mat to converting .acq to .mat and load it to matlab.
.acq has many versions, and the load_acq matlab code seems not working >3.9 (current one is 4.1 and 5.0).
Please check below discussions.
https://sourceforge.net/p/pspm/discussion/bugs/thread/b0a1bdbc/
Below is the .acq file I attached : )
As far as I know, the only working solution for Biopac .acq is using acq2mat to converting .acq to .mat and load it to matlab.
acq2mat is based on load_acq, which is the same function as the one I used in Brainstorm.
Therefore it can't be working for your file.
Have you managed to convert successfully your files with this function previously?
Please check below discussions.
It looks like the most up-to-date solutions are based on bioread:
This is a Python library, I won't be able to reuse it directly and won't have time to recode it in Matlab.
Can you try converting your .acq files to .mat, then open and plot them and in Matlab to make sure it works better than load_acq?
Then share the .mat files here, and I will edit the BIOPAC reader so that you can easily import them into Brainstorm.