Hello dear Brainstorm community,
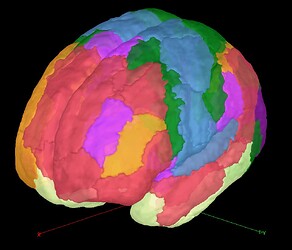
I would like to know if there is a simple way to automatically assign dipoles from an already existing source grid to brain regions (e.g., DorsAttn_FEF_1 L/R, ... from the Schaefer2018_100_7net surface atlas, shown below, for example)?
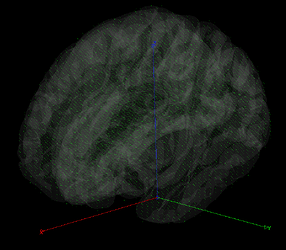
My source space consists of 13430 unconstrained dipoles distributed over a 5mm resolution 3D grid (shown below). I generated my headmodel with DUNEuro from the standard 1T ICBM152 MRI from Brainstorm's default anatomy, a 3-layer 12841V FEM mesh (iso2mesh-2021, MergeMesh MaxVol=1) and a 64-electrode Biosemi headcap.
I read the Source estimation (https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation), the Volume source estimation (https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource), the Scouts (https://neuroimage.usc.edu/brainstorm/Tutorials/Scouts) as well as the DBA (https://neuroimage.usc.edu/brainstorm/Tutorials/DeepAtlas) tutorials. I also visited several discussions on this forum, but I still struggle to generate the same 13430V source grid, as just above, with its dipoles allocated to the brain regions of, for instance, the Schaefer2018_100_7net surface atlas.
Similarly to the DBA tutorial, I generated the source grid just above from a surface file called "cortex_mixed_schaefer2018_100" which is the result of merging the default "cortex_15002V" surface file with a "schaefer2018_100_7net (mni-segment)" surface file. The latter is directly generated from the Schaefer2018_100_7net MNI parcellation of the ICBM152 MRI file.
When computing the headmodel, I checked the "Custom source model" option.
Unfortunately, none of the 18597 dipoles from the source grid just above matches with any of the 13430 dipoles from my initial source grid.
I hope I made myself understood. Please feel free to correct me as I might have misunderstood several key points from the tutorials.
Best,
Valentin