Dear users,
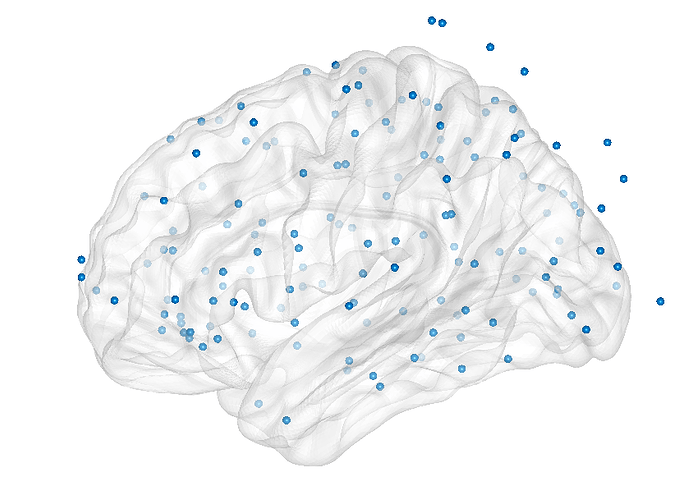
I am trying to convert the seeds coordinates from the Destrieux atlas to the MNI reference. However, there seems to be a discrepancy when plotting the obtained points on NBS or BrainNet viewer (as shown in the picture), as the points are beyond the limit of the surface.
I am using the ICBM152 template and the cs_convert function from SCS to MNI.
I hope you may help me to figure out what I am doing wrong.
Many thanks in advance for your help!
I'm sorry, the information you provide is not sufficient to guess what is going wrong in your procedure.
The MNI normalization used in Brainstorm is an approximation using only linear 4x4 transformation between the subject and and MNI152 brains. It is not an accurate procedure, and performing an MNI normalization with a different algorithm in a different program could lead to different results. That said, the points at the top look far off, so there is probably an other additional problem. It looks like your cloud of blue points is not aligned with the surface... There are many steps in this conversion and you have to check each one of them.
Maybe start by following the conversion of one single point in a region easy to identify both the subject MRI and in the MNI template.
Note that the seeds of the Destrieux atlas are not always meaningful. The seed of a scout can be placed anywhere in this scout... If you used the center of mass of the scout, you might get something a bit more meaningfull. But a more general recommendation is that if you want to work in volume and use volume-based registrations, work with volume source models and volume scouts instead.
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource
If you work at the surface level (surface source models, surface scouts), use the surface-based registration tools. If you have processed the MRI of your subject with FreeSurfer of BrainSuite, you can project the scouts from the individual anatomy to the MNI brain in a much more reliable way.
https://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects
Dear Francois,
Many thanks for your help. In fact, I am aware of the fact that the seeds might not be very meaningful, as I was using them just to have a rough idea of the network topography. However, I expected them at least not to exceed the surface limit, as in the SCS system those coordinates are points of the surface itself.
My procedure is simply to run this line:
seeds_mni=cs_convert(sMRI,'scs','mni',seeds);
Where sMRI is the exported MRI file from the ICBM152 template. seeds is obtained from the surface file of the same template. I also make sure that seeds is in meters, and convert seeds_mni in millimeters for plotting.
If you work at the surface level (surface source models, surface scouts), use the surface-based registration tools. If you have processed the MRI of your subject with FreeSurfer of BrainSuite, you can project the scouts from the individual anatomy to the MNI brain in a much more reliable way.
https://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects
I will definitely look at this, thank you very much.
Ramtin.
No, forget about this... I though you were working on an individual subject. My previous post was completely missing your point.
Then there is something wrong in the way you manipulate the coordinates, as the MNI coordinates you get from Brainstorm should be correct. This IS the MNI ICBM152 template, there is no possible problem of registration to the MNI template.
Use the MRI viewer and navigate in the volume to make sure that what you get out of cs_convert is correct. If you think there is a problem with the Brainstorm MRI viewer, you can also try get the MNI coordinates of some specific structures with another program, like MRIcron.
In principle, I am working with individual subjects, therefore this chapter will be useful for me.
In this specific case (the one I opened the post for) I am testing the MNI conversion using the template, which is why I would not expect this kind of registration errors, as you just stated.
To save these coordinates I still need to go through the cs_convert step, as the coordinates are saved as SCS in the surface file, is that correct?
Thank you.
To save these coordinates I still need to go through the cs_convert step, as the coordinates are saved as SCS in the surface file, is that correct?
Correct.
But it doesn't call any additional registration procedure, it should just returns the corresponding coordinates.
Dear Francois,
It seems that the problem is sorted. When exporting the surface template variable, Matlab was not replacing a subject specific one I previously saved in the workspace. Awkward mistake, I am sorry. Now the registration looks reasonable.
I really appreciate your help.
Ramtin
EDIT: reducing the size of the nodes in the plot, some of them are still a little floating. I will explore further, but the outcome is reasonable now.