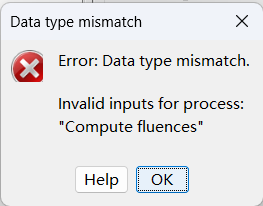
Hi, I tried to analysis my fNIRS data on the cortical surface, when I running compute fluences with raw d OD signal, an error "Data type mismatch" emerged. How to solve this problem?
Should I use averaged Hb? thanks
Hi @woshibeauty
Can you post the entire error message that you are obtaining?
@edelaire can you please check it?
Hello,
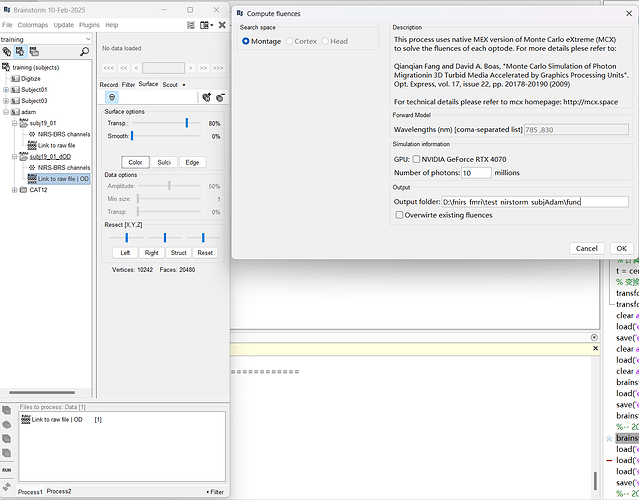
You are trying to do the fluence on a 'link to raw' type of file. This is not suppoorted yet.
You need to import the file in the database, using right click > import in database:
i will try to modify the process this week to have it work also for link to raw file.
Edit2: don't forget to select your GPU in the fluence panel.
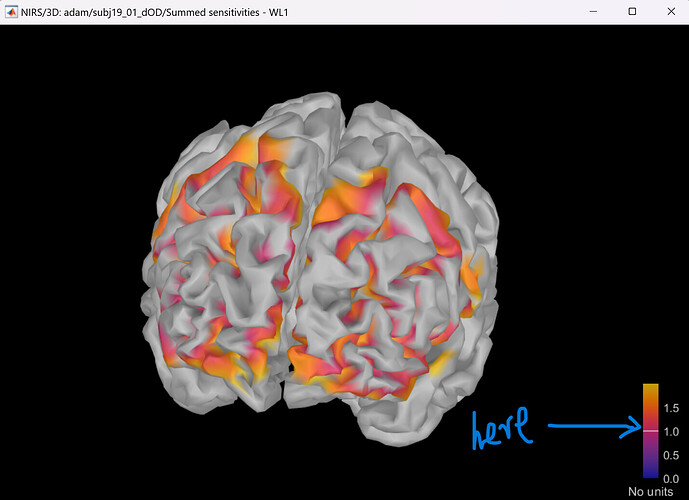
Thank you Delaire, I have successfully got the sensitivity map.
A further question: why the middle value(=1, in the attached figure) always have the same color as unmeaseured surface in all my figures, this makes the pictures weird. Do you have any suggestions to solve this problem?
The white line in the colorbar indicates that there is a threshold applied, and anything below it ( <1) will not be shown (therefor gray). To change, just set the Amplitude slider in the Surface tab to 0%
https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Display:_Cortex_surface
Thank you all for your help.
May I ask one more question? @edelaire In your previous tutorial and its online video, you imported fMRI map into nirstorm? I would like to ask how to do that? Since I want to analysis my simultaneous fNIRS/fMRI data.
Just to be sure, when you say you want to import fMRI data in Brainstorm; do you mean you want to project a BOLD tomecourse on the cortex; or import a static volume (for example, the result of a GLM analysis) that will again be projected on the cortex ?
Yes, I would like to import the later one.
And it will be processed in SPM.
Hi @edelaire,
I have attempted to resolve this on my own, but I am still unable to find a solution. Your guidance on how to import fMRI processed data into NIRSTORM would be greatly appreciated.
Thank you in advance!
Hi.
Sorry for the delay. I just finished writing the tutorial part on that here: https://neuroimage.usc.edu/brainstorm/Tutorials/NIRS_Optimal_montage#Personalized_optimal_montage
You can especially read the section " Projecting a volume onto the cortical surface"
Let me know if there is anything unclear so I can improve the tutorial