Hello everyone, i hope that you are doing well !
I'm using two devices from Artinis MS, Brite MKII and Brite MKIII.
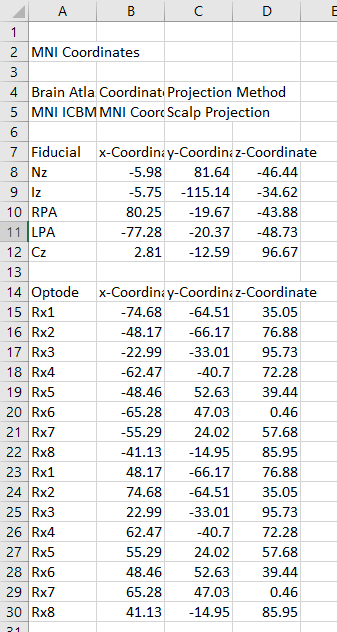
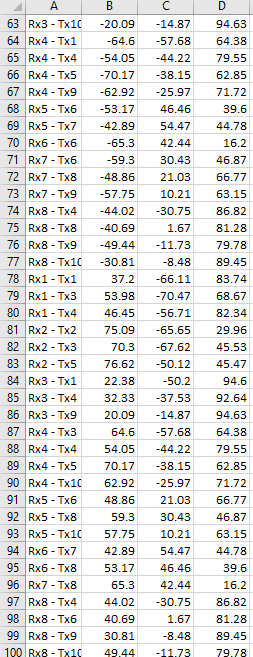
In order to have a good look of my channels on the scalp (and with the future idea of generating a sensitivity maap), i used a Polhemus Fastrack to digitize my optodes. This was done in the Oxysoft software provided by Artinis MS. This results in a .xlsx file which look like this :
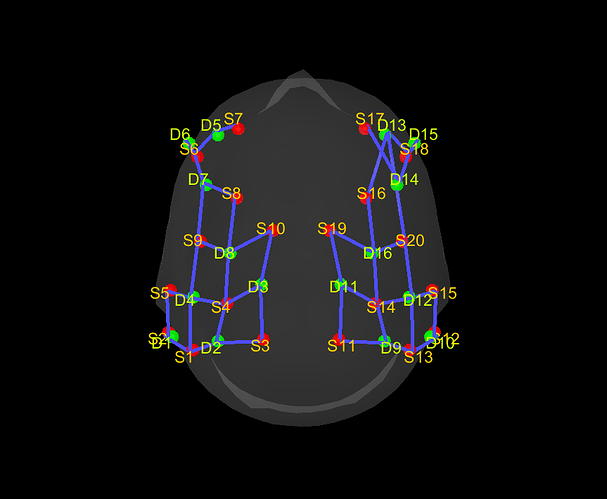
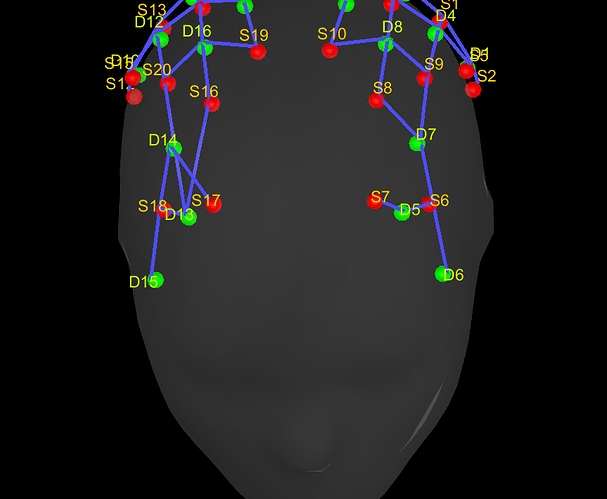
Then, i would like to import these coordinates in NIRSTORM using the Colin27 4 NIRS template (or another if preferable) but i can't find a way to do it. So i tried to manually change it with "Edit channel file", that works but does not fit the MRI template. I can't provide a screenshot because I deleted it by accident but i saved it in a .pos file (but when i try to import this file with "Add EEG positions", it does not change the coordinates) :
channel.pos (2 Bytes)
I also found a similar discussion but it had no solution : How to import the MNI probe coordinate of NIRS into brainstorm?
What should I do ? Is there a tutorial that I missed or that I did not understand ? In the NIRS finger tapping tutorial, the coordinates are provided with Brainsight but both fiducials and optodes txt are not "written" as mines.
Thank you very much for your help,
Kind regards,
Brieuc