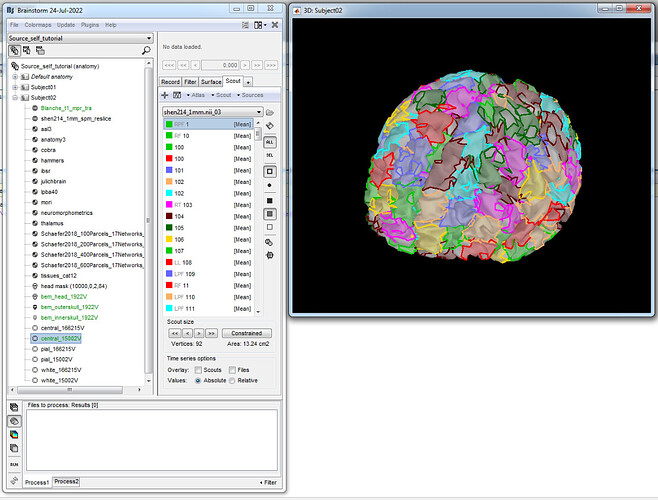
I have a .nii file with an atlas that has 214 ROIs, each one coded by intensity level. When I open it with the MATLAB function niftiread I see that it has 215 unique values (0 to 214).
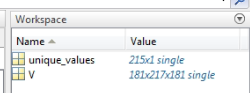
I was able to load it into my cortex file after segmentation of the subject's MRI, and it looks like this:
However, when I downsample to this atlas after source reconstruction, I get 216 time series. How can this be the case? (The example corresponds to one trial with 2500 temporal samples).

Thanks in advance.
The step you need to check is the import of your parcellation.
Check the Atlas field in the cortex surface:
https://neuroimage.usc.edu/brainstorm/Tutorials/Scouts#On_the_hard_drive
Note that we don't recommend using this process "Downsample to atlas".
I added a new warning in this process:
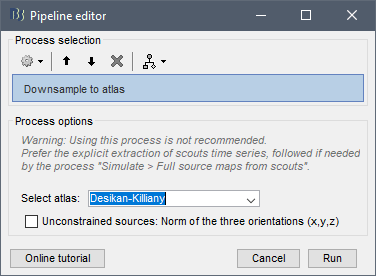
When I'm loading the atlas it says it's creating 214 scouts:
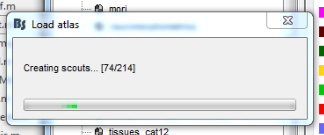
But in the end it creates 216, I don't really understand? The atlas field in the cortex surface has 216 regions.
Note that using a volume atlas from a .nii volume to create surface atlases is not an accurate procedure.
If you need to use an volume anatomical atlas with volume ROIs, it could be a better option to use volume source models and volume scouts.
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource
If you want to keep working in the same direction, please share the example file you are trying to load here: upload it (zipped) somewhere and post the download link here.
For reproducing the behavior you describe, I would also need:
- the cortex surface on which you are trying to load the atlas an (the .mat file from the Brainstorm database)
- the reference MRI of the subject (the first one in the anatomy folder)
Please zip these two files together with the .nii atlas and share it with us.

![]()