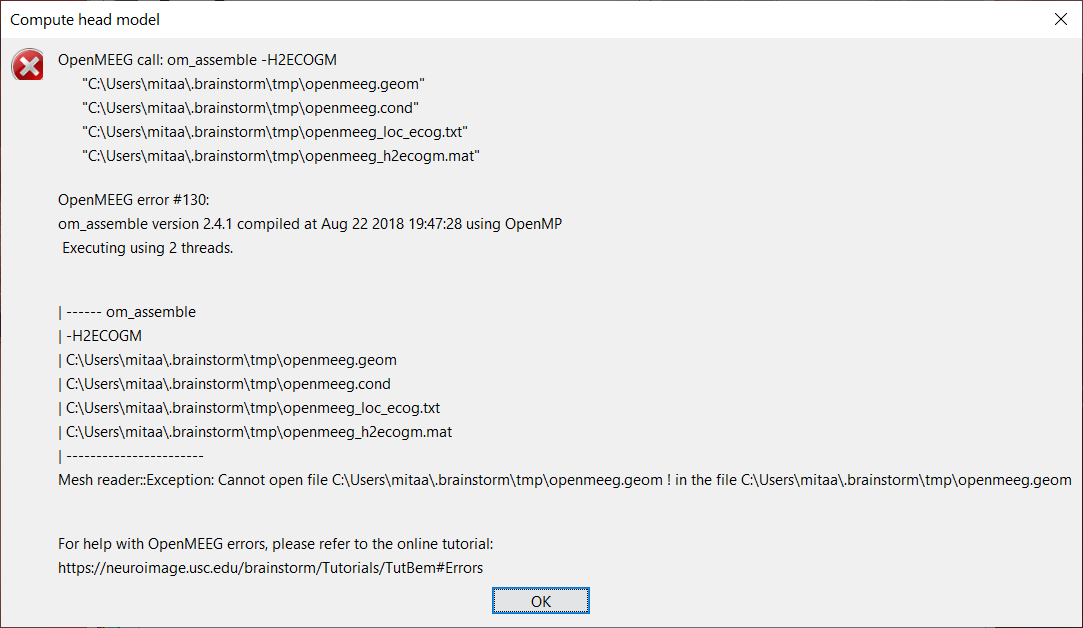
Hi, I have a problem with calculating the BEM volume. As settings, I select MRI volume and in the OpenMEEG options "Use adaptative integration".
As soon as I give the OK, in the Command Windows various errors begin to appear that I have reported below. The version of the toolbox I use is 2.4.1 because the latest version didn't even allow me to do the BEM Cortex calculation. At least with this version, I can calculate the BEM Cortex model.
I await your news.
Index exceeds the number of array elements (0).
Error in cs_compute (line 158)
AC(2) PC(2) IH(2);
Error in tess_envelope (line 171)
Transf = cs_compute(sMri, 'tal');
Error in panel_sourcegrid>CreatePanel (line 61)
[sEnvelope, sCortex] = tess_envelope(CortexFile, 'convhull',
GridOptions.nVerticesInit, .001, []);
Error in panel_sourcegrid (line 30)
eval(macro_method);
Error in gui_show_dialog (line 51)
[bstPanel, panelName] = fcnPanel('CreatePanel', varargin{:});
Error in bst_headmodeler (line 256)
sGrid = gui_show_dialog('Volume source grid', @panel_sourcegrid, 0, [],
OPTIONS.CortexFile);
Error in panel_headmodel>ComputeHeadModel (line 669)
[OPTIONS, errMessage] = bst_headmodeler(OPTIONS);
Error in panel_headmodel (line 27)
eval(macro_method);
Error in panel_protocols>TreeHeadModel (line 1251)
[OutputFiles, errMessage] = panel_headmodel('ComputeHeadModel', iChanStudies);
Error in panel_protocols (line 44)
eval(macro_method);
Error in tree_callbacks>@(h,ev)panel_protocols('TreeHeadModel',bstNodes) (line 2505)
gui_component('MenuItem', jPopup, [], 'Compute head model',
IconLoader.ICON_HEADMODEL, [], @(h,ev)panel_protocols('TreeHeadModel', bstNodes));