Dear BS community,
I am using BS for source localization for EEG data. I was able to follow the guidelines of several tutorials in order to estimate the TF representation (using Morlet wavelet) on source data for full cortical maps.
Description of the dataset:
EEG data collected from 32 subjects, where we had three motor imagery conditions. Each subject performed N trials of motor imagery for each of the conditions, each lasting three seconds. We are interested in time-frequency analysis at the source level. Data was pre-processed using EEGLAB.
Here are the steps I followed:
For each condition:
- Downsample to 200Hz from 1000 Hz.
- Compute covariance matrix (pre-stim as baseline). Estimate the source for each trial using MNE (output is in pA.m)
- Compute TF maps for each trial (optimized for faster computation)
• Avergae across frequency bands
• Average every 50ms timebin - Average TF maps across trials within a subject.
- Perform normalization for the averaged map using db method (10log10(activity/baseline)).
- Average maps across subjects.
- Plot the average time-frequency map.
I am able to see consistency between cortical maps and sensor level topographies for my frequency bands of interest.
My question:
- I would like to conduct statistical testing to compare conditions pairwise. Since I have the time dimension and the spatial dimension, the number of comparisons is huge. Reading through the documentation, I realized I can use cluster based permutation testing.
What I tried already:
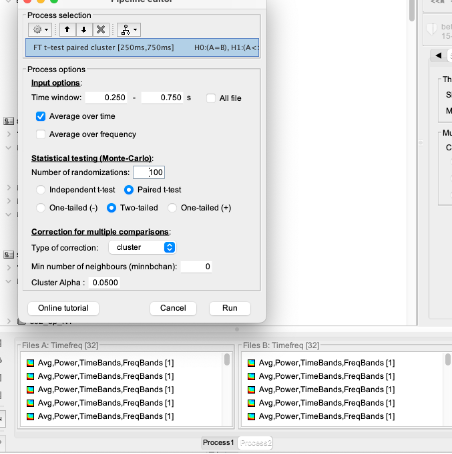
- I tried to use process 2 to compare TF cortical maps for two of my conditions.
I placed the 32 files (TF cortical maps) for each condition in one pane.
-
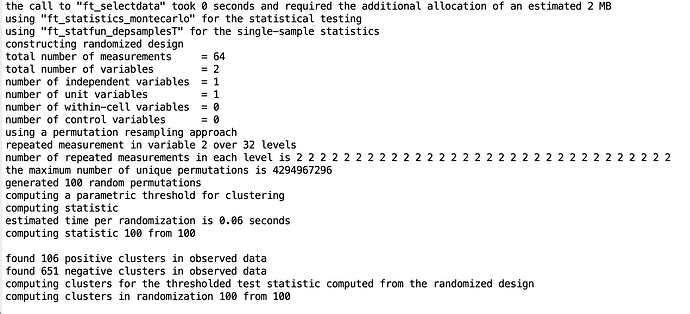
The process runs smoothly:
-
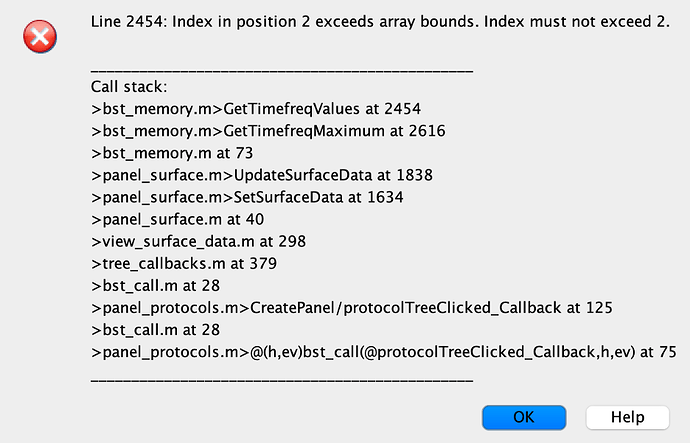
However, I get the error below when I try to load the outcome: