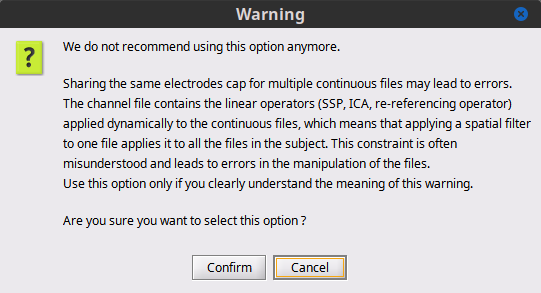
Hello BSt support:
I am relearning how to set up a processing pipeline. I was able to create a sequence as follows: baseline removal, bandpass filter, notch filter, and resample. I saved this pipeline, then ran it on a single subject's raw EEG file. It created 4 new nodes, each with a further processing step in the node name. I looked at the EEG from each of these 4 nodes and they looked correct.
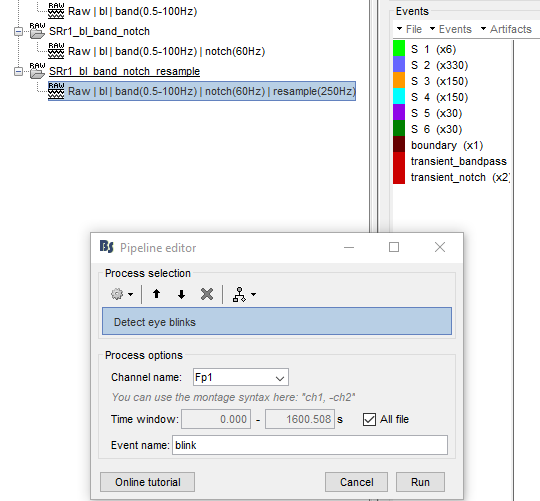
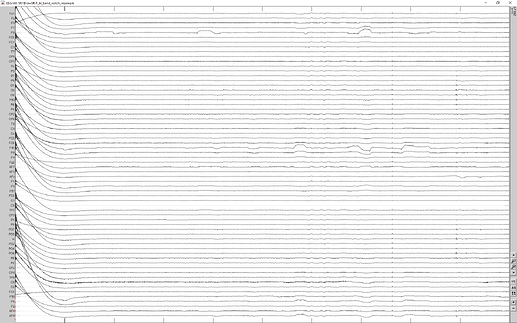
Then I cleared the Process 1 box and dragged the last node to it. My intent was to operate only on this last node. I read up on the blink removal, and manually ran the two steps needed from the Event box artifact tab: detect eye blinks and SSP EOG blink. Then I looked at all 5 nodes and ALL 5 of them had had their blinks removed. This does not seem right to me.
Here are some screen shots:
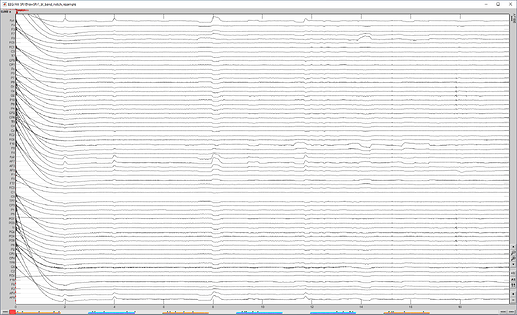
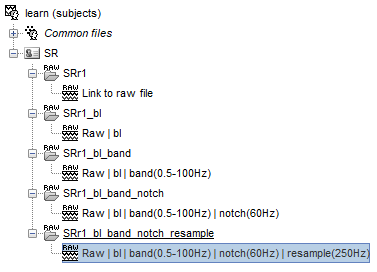
after running saved pipeline (bandpass, notch, resample)
some steps:
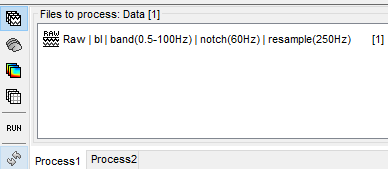


after blink removal:
I also went back and checked the first step of the pipeline, DC removal, and it had not been performed. I thought it had but I was using the Dc remove display option the first time I looked at it.
Maybe I do not fully understand the link to file aspect of all this?
Finally, I was confused by the Projector component screen - where is this documented?