Dear Raymundo,
Sorry to bother you again  . After importing the volume atlas from the subject anatomy as described before, I then tried to extact the volume scouts using the command lines. The script looks like the following:
. After importing the volume atlas from the subject anatomy as described before, I then tried to extact the volume scouts using the command lines. The script looks like the following:
sFiles = bst_process('CallProcess', 'process_extract_scout', sFiles, [], ...
'timewindow', [0, 1.998], ...
'scouts', {'Volume 9557: aal3', {'Left Amygdala', 'Left Angular gyrus', 'Left Anterior cingulate cortex-pregenual', 'Left Anterior cingulate cortex-subgenual', 'Left Anterior cingulate cortex-supracallosal', 'Left Anterior orbital gyrus', 'Left Calcarine fissure and surrounding cortex', 'Left Caudate nucleus', 'Left Crus I of cerebellar hemisphere', 'Left Cuneus', 'Left Fusiform gyrus', 'Left Gyrus rectus', 'Left Heschls gyrus', 'Left Hippocampus', 'Left IFG pars orbitalis', 'Left Inferior frontal gyrus-opercular part', 'Left Inferior frontal gyrus-triangular part', 'Left Inferior occipital gyrus', 'Left Inferior parietal gyrus-excluding supramarginal and angular gyri', 'Left Inferior temporal gyrus', 'Left Insula', 'Left Intralaminar', 'Left Lateral geniculate', 'Left Lateral orbital gyrus', 'Left Lateral posterior', 'Left Lenticular nucleus-Pallidum', 'Left Lenticular nucleus-Putamen', 'Left Lingual gyrus', 'Left Lobule VI of cerebellar hemisphere', 'Left Medial Geniculate', 'Left Medial orbital gyrus', 'Left Mediodorsal lateral parvocellular', 'Left Mediodorsal medial magnocellular', 'Left Middle cingulate & paracingulate gyri', 'Left Middle frontal gyrus', 'Left Middle occipital gyrus', 'Left Middle temporal gyrus', 'Left Nucleus accumbens', 'Left Olfactory cortex', 'Left Paracentral lobule', 'Left Parahippocampal gyrus', 'Left Postcentral gyrus', 'Left Posterior cingulate gyrus', 'Left Posterior orbital gyrus', 'Left Precentral gyrus', 'Left Precuneus', 'Left Pulvinar anterior', 'Left Pulvinar inferior', 'Left Pulvinar lateral', 'Left Pulvinar medial', 'Left Rolandic operculum', 'Left Superior frontal gyrus-dorsolateral', 'Left Superior frontal gyrus-medial', 'Left Superior frontal gyrus-medial orbital', 'Left Superior occipital gyrus', 'Left Superior parietal gyrus', 'Left Superior temporal gyrus', 'Left Supplementary motor area', 'Left SupraMarginal gyrus', 'Left Temporal pole: middle temporal gyrus', 'Left Temporal pole: superior temporal gyrus', 'Left Thalamus-Anteroventral Nucleus', 'Left Ventral anterior', 'Left Ventral lateral', 'Left Ventral posterolateral', 'Right Amygdala', 'Right Angular gyrus', 'Right Anterior cingulate cortex-pregenual', 'Right Anterior cingulate cortex-subgenual', 'Right Anterior cingulate cortex-supracallosal', 'Right Anterior orbital gyrus', 'Right Calcarine fissure and surrounding cortex', 'Right Caudate nucleus', 'Right Crus I of cerebellar hemisphere', 'Right Cuneus', 'Right Fusiform gyrus', 'Right Gyrus rectus', 'Right Heschls gyrus', 'Right Hippocampus', 'Right IFG pars orbitalis', 'Right Inferior frontal gyrus-opercular part', 'Right Inferior frontal gyrus-triangular part', 'Right Inferior occipital gyrus', 'Right Inferior parietal gyrus-excluding supramarginal and angular gyri', 'Right Inferior temporal gyrus', 'Right Insula', 'Right Intralaminar', 'Right Lateral geniculate', 'Right Lateral orbital gyrus', 'Right Lateral posterior', 'Right Lenticular nucleus-Pallidum', 'Right Lenticular nucleus-Putamen', 'Right Lingual gyrus', 'Right Lobule IV-V of cerebellar hemisphere', 'Right Lobule VI of cerebellar hemisphere', 'Right Medial Geniculate', 'Right Medial orbital gyrus', 'Right Mediodorsal lateral parvocellular', 'Right Mediodorsal medial magnocellular', 'Right Middle cingulate & paracingulate gyri', 'Right Middle frontal gyrus', 'Right Middle occipital gyrus', 'Right Middle temporal gyrus', 'Right Nucleus accumbens', 'Right Olfactory cortex', 'Right Paracentral lobule', 'Right Parahippocampal gyrus', 'Right Postcentral gyrus', 'Right Posterior cingulate gyrus', 'Right Posterior orbital gyrus', 'Right Precentral gyrus', 'Right Precuneus', 'Right Pulvinar anterior', 'Right Pulvinar inferior', 'Right Pulvinar lateral', 'Right Pulvinar medial', 'Right Rolandic operculum', 'Right Superior frontal gyrus-dorsolateral', 'Right Superior frontal gyrus-medial', 'Right Superior frontal gyrus-medial orbital', 'Right Superior occipital gyrus', 'Right Superior parietal gyrus', 'Right Superior temporal gyrus', 'Right Supplementary motor area', 'Right SupraMarginal gyrus', 'Right Temporal pole: middle temporal gyrus', 'Right Temporal pole: superior temporal gyrus', 'Right Thalamus-Anteroventral Nucleus', 'Right Ventral anterior', 'Right Ventral lateral', 'Right Ventral posterolateral'}}, ...
'flatten', 0, ...
'scoutfunc', 'mean', ... % Mean
'pcaedit', struct(...
'Method', 'pca', ...
'Baseline', [-0.1, 0], ...
'DataTimeWindow', [0, 1], ...
'RemoveDcOffset', 'file'), ...
'isflip', 1, ...
'isnorm', 0, ...
'concatenate', 1, ...
'save', 1, ...
'addrowcomment', 1, ...
'addfilecomment', []);
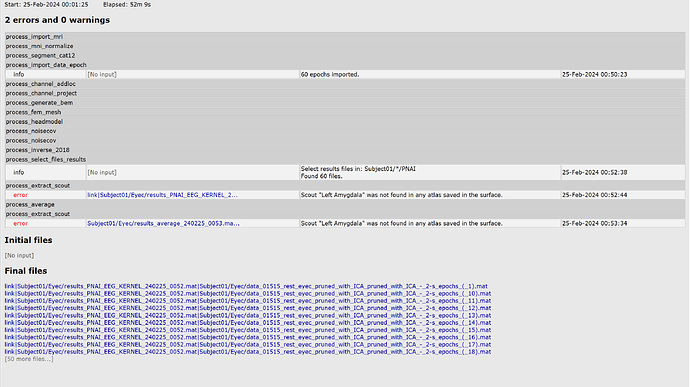
The scout labels are just the same as those showed in the GUI. However, when I run the script, an error occurred as below:
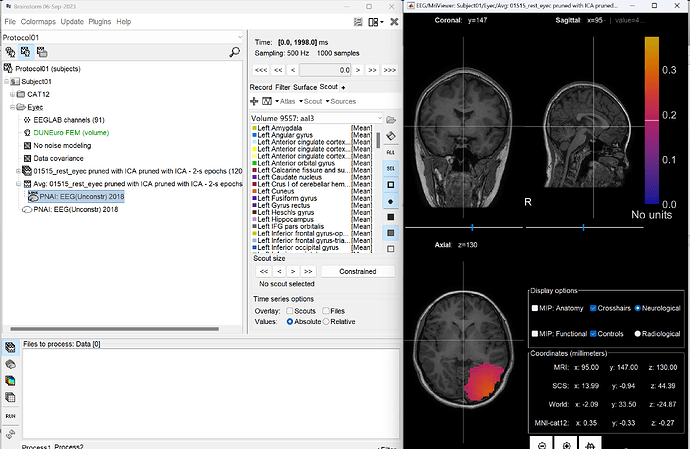
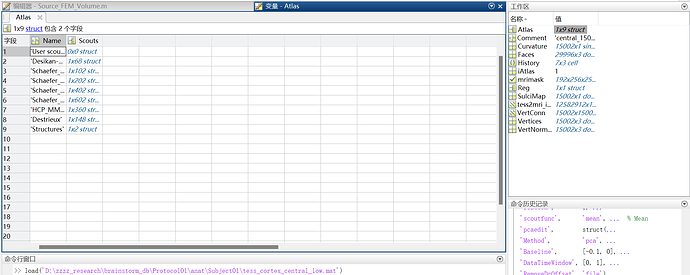
I tried to extract the scout sources from the epoch and average files. It seems the volume atlas should have be saved in the surface to be detected?
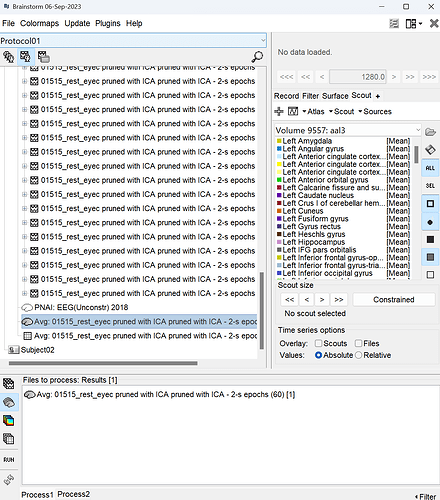
When I did the same process in the GUI, It work successfully and generated the corresponding scout source file.
I just cann't figure out what caused the error. Should I attached the volume atlas to the surface file? If so, what can I do in the script  ? Use the panel_scout('SaveScouts') function?
? Use the panel_scout('SaveScouts') function?
Best,
Greatson Wu