Hi
I'm trying the scripting function for the first time to automate most of my pre-processing. I've no prior experience with coding in Matlab/Brainstorm. I have 60 subjects, one EEG acquisition one per subject. I tried entering the below script which I found in another thread on this forum to create multiple subjects. Under a new protocol, I selected file > command window and copied pasted the below script in MATLAB command window (I put in a number for 'n' in first line, also tried for 'no' in first and second line). I got the below answer in the cmd.exe window. Could you advise what the right steps are to run this script?
function create_nSubjects(start, noSubject)
range = start:noSubject;
for i = range
if i < 10
subjectname = [‘Subject’, num2str(0), num2str(i)];
else
subjectname = [‘Subject’, num2str(i)];
end
[sSubject, iSubject] = db_add_subject(subjectname);
end
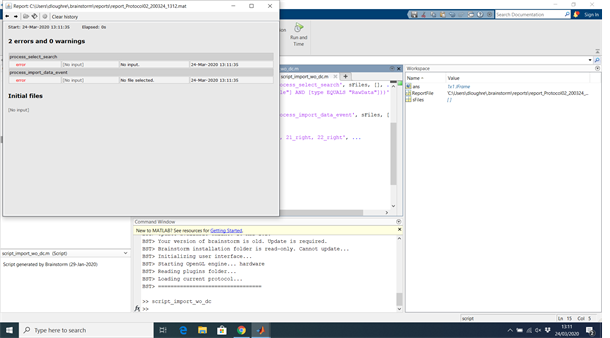
BST> Error executing command:
Error using panel_command>CreatePanel/ButtonRun_Callback (line 66)
Error: Function definitions are not permitted in this context.