Hi Martin,
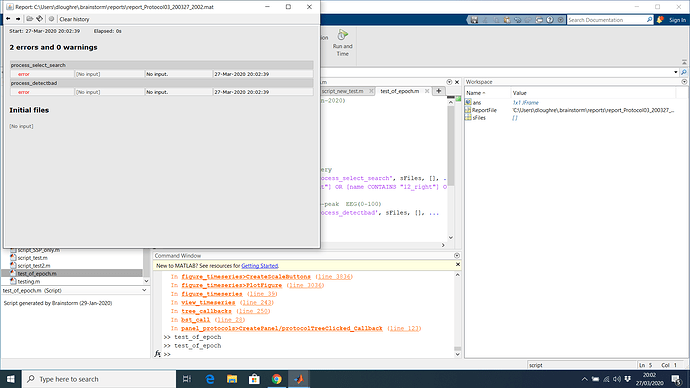
I also tried the below script using the tag process but this didn't work either. I can run the process accurately if I copy the entire protocol into the run process box and generate a script listing all the database files and then use the search function within this script. It will be useful to know if there's another way to do it though rather than have every file in the database listed in the script.
% Script generated by Brainstorm (29-Jan-2020)
% Input files
sFiles = [];
% Start a new report
bst_report('Start', sFiles);
% Process: Select file names with tag: resample
sFiles = bst_process('CallProcess', 'process_select_tag', sFiles, [], ...
'tag', 'resample', ...
'search', 2, ... % Search the file names
'select', 1); % Select only the files with the tag
% Process: Import MEG/EEG: Events
sFiles = bst_process('CallProcess', 'process_import_data_event', sFiles, [], ...
'subjectname', '', ...
'condition', '', ...
'eventname', '11_right, 12_right, 21_right, 22_right', ...
'timewindow', [], ...
'epochtime', [-0.1992, 1], ...
'createcond', 1, ...
'ignoreshort', 1, ...
'usectfcomp', 1, ...
'usessp', 1, ...
'freq', [], ...
'baseline', []);
% Save and display report
ReportFile = bst_report('Save', sFiles);
bst_report('Open', ReportFile);
% bst_report('Export', ReportFile, ExportDir);