I have import the MRI and CT scan in brainstorm and I edit all SEEG contacts in the MRI according to the tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity
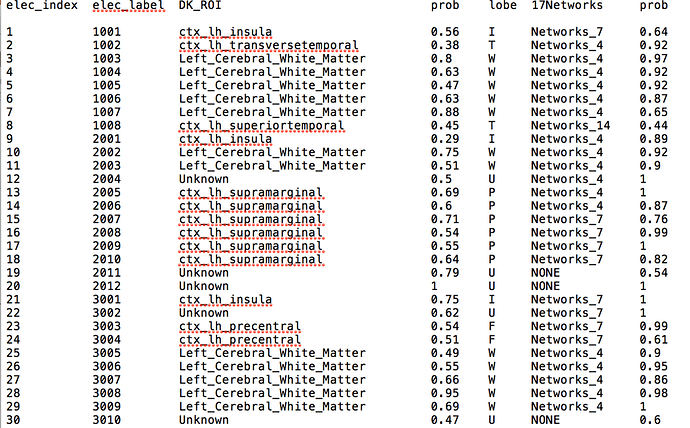
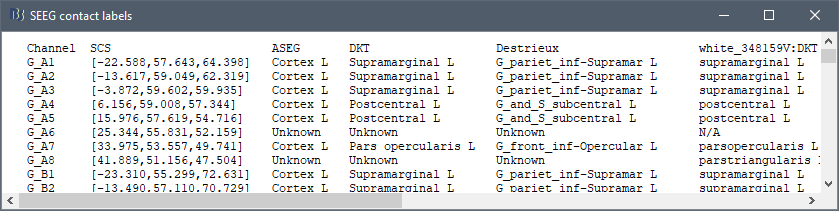
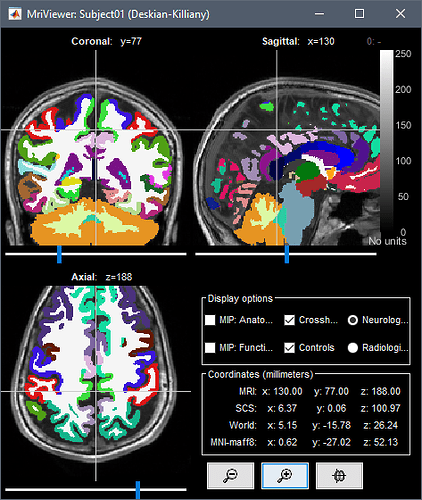
Whether I can use Brainstorm to tell anatomical location of each contacts like this:
No, Brainstorm cannot produce (yet) these kinds of lists.
This will be added at some point, but not immediately.
You would need to do it manually, probably from a volume MNI atlas.
This is the goal of the IntrAnat software, but unfortunately this is very complicated to install:
https://github.com/ftadel/IntrAnat/wiki
Ok,I will have a try! Thanks!
Have you solved this problem? I recently tried to use the tools provided by Fieldtrip, but the accuracy of the results is not very good
I will most likely work on this starting from next week.
Thank you for your patience.
Thanks for your reply!We are looking forward to this new feature, it will be very helpful to our current work
Done!
Thanks a lot! You are so efficient!
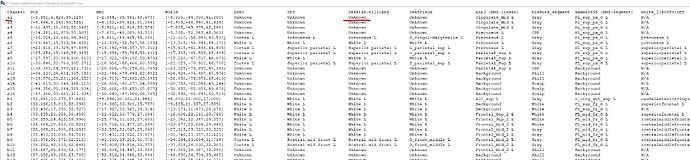
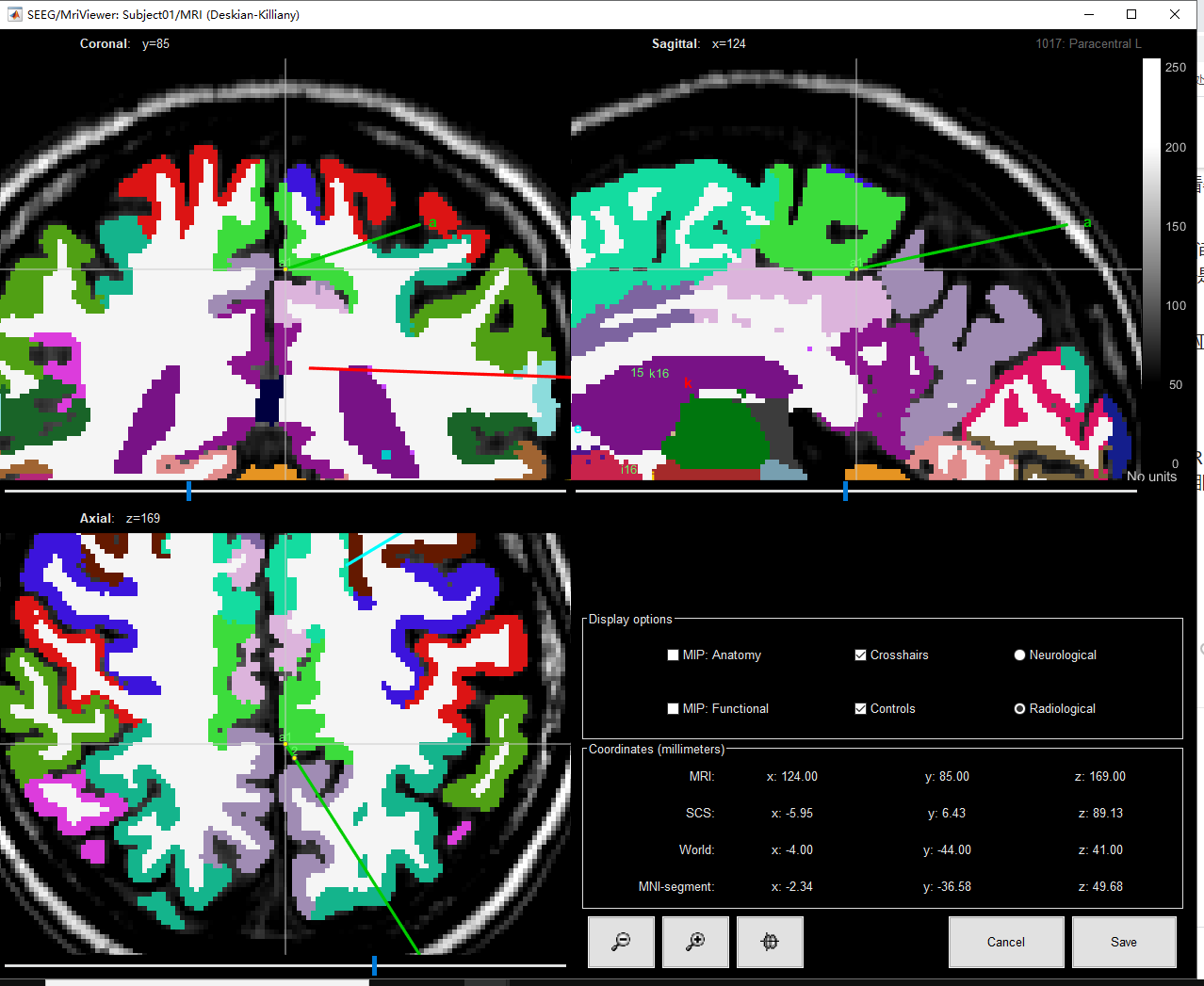
But in the process of trying, I found that although AAL and other atlas based on MNI coordinates are normal, there are many contacts marked as 'unknown' in some other atlas. In addition to the contacts that are outside the skull, there are some contacts that can display the region normally in the MRI viewer, are also marked as 'unknown', which means that the table and the MRI viewer have different results for the same location. What is the problem?
In your example, it does look like your A1 contact is not in any DK label.
The FreeSurfer cortex parcels are not inflated much in volume, therefore there are a lot of "Unknown" voxels in the sulci and the interhemispheric fissure.
I guess you used a sphere with a small radius, like 1mm?
How many voxels does it say its using (see the Matlab command window)?
If you increase the radius (>= 3mm), do you still see all Unknown values you don't expect?
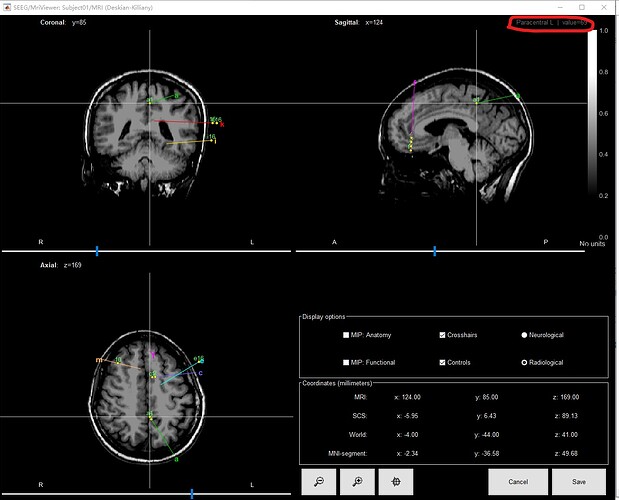
The concerning part is that the closest voxel to A1 are different in the MRI viewer and in the exported table. There might be something wrong here, but I'm not sure how to debug this...
Is the "Paracentral L" information from the MRI viewer really coming from the same Desikan-Killiany volume that is used in the computation of the labels?
(right-click on the MRI viewer > Anatomical atlas > ...)
I think I could reproduce what you mentioned. There was a discrepancy between 1) the voxel selected in the MRI viewer when clicking on an electrode in the iEEG tab, and 2) the voxel identified as the closest voxel to the contact when creating the table of labels.
I noticed that there could be very small differences (~0.2mm) between the tip of the electrode (which corresponds to the center of a voxel) and the position of the first contact in the channel file. This difference is almost invisible at the level of precision manipulated in the Brainstorm MRI viewer, but can cause the rounding of the coordinates to an integer number of voxel to jump to a different voxel.
It is not very meaningful per se, as we have to understand that with these tools, we can't be achieve a submillimetric precision, but it leads to very counter-intuitive observations, as you reported.
I tried to address this issue with this commit: https://github.com/brainstorm-tools/brainstorm3/commit/4f55e8af11f36e3371cacd0dd8d3e0588a296641
Now, when clicking on an electrode in the iEEG panel, it should jump to the position of the first contact on the electrode in the MRI viewer, instead of jumping to the tip of the electrode. You should now always get the same label at the top-right corner of the MRI viewer and in the labels table (when using a sphere radius <= 1).
Update Brainstorm to get this fix.
Does it explain what you reported?
Thank you for your reply. I carefully observed the problematic contacts and invited the neurosurgery experts in our group to participate in the discussion. Such a contact is indeed at the edge of the DK partition.
In some other software (such as fieldship), such points will still be assigned to the closest area, and a percentage will be indicated to indicate the percentage of the area in the sphere, even if the percentage is less than 50%. I guess that in our new brainstorm feature, when most of the voxels in the sphere are not in any partition, they will be marked as "N/A", right? I didn't realize this problem at first
As for the problem of the different areas of the MRI viewer and the contact in the list, the radius of the sphere in the screenshot I gave is set to the default 3mm. In the new update, unfortunately this problem still exists, but your answer made me aware that this is probably because the contact is located at the edge of the area.
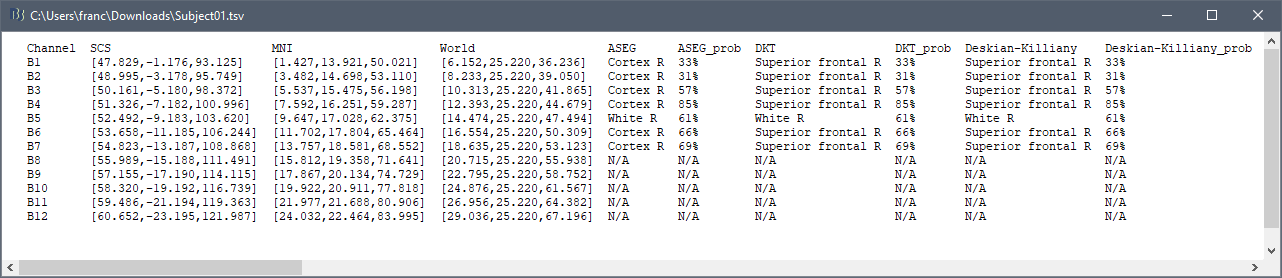
Can you provide more format export versions of this table? Such as excel or .txt? Thank you very much for your efficient update, which has provided a lot of help for our research
I guess that in our new brainstorm feature, when most of the voxels in the sphere are not in any partition, they will be marked as "N/A", right?
No, if there is only one non-zero voxel in the sphere, the contact should be attributed a label.
(if this is not the case, make sure you really updated Brainstorm to include my last modifications)
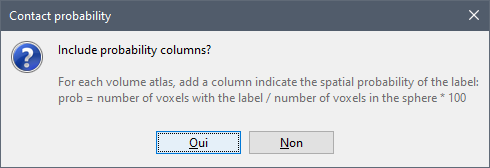
a percentage will be indicated to indicate the percentage of the area in the sphere
Do you think you would use routinely the % of the label within the sphere if you had this information available in the table?
How would you recommend making this information accessible? An extra adjacent column for each column with the spatial probability (number of voxels with the label / number of voxels in the sphere)? Or include this % value in the label?
Can you provide more format export versions of this table? Such as excel or .txt?
The .tsv file is a text file that you can:
- read with any text viewer
- open direclty with Excel
Nice work! Thank you very much!
I am very sorry that I did not reply to you before. I have been busy with an academic conference some time ago.
I just performed some simple tests, the function is very complete, it is very helpful to our current research, and it is expected that our laboratory will gradually transition from using fieldtrip to using brainstorm.
Thank you for your efficient update! Hope we can keep in touch like this all the time!